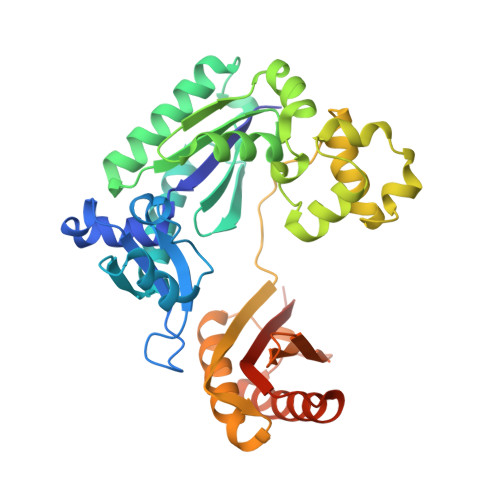
Structure and Activity of Y-Class DNA Polymerase Dpo4 from Sulfolobus Solfataricus with Templates Containing the Hydrophobic Thymine Analog 2,4- Difluorotoluene.
Irimia, A., Eoff, R.L., Pallan, P.S., Guengerich, F.P., Egli, M.(2007) J Biological Chem 282: 36421
- PubMed: 17951245
- DOI: https://doi.org/10.1074/jbc.M707267200
- Primary Citation of Related Structures:
2V9W, 2VA2, 2VA3 - PubMed Abstract:
The 2,4-difluorotoluene (DFT) analog of thymine has been used extensively to probe the relative importance of shape and hydrogen bonding for correct nucleotide insertion by DNA polymerases. As far as high fidelity (A-class) polymerases are concerned, shape is considered by some as key to incorporation of A(T) opposite T(A) and G(C) opposite C(G). We have carried out a detailed kinetic analysis of in vitro primer extension opposite DFT-containing templates by the trans-lesion (Y-class) DNA polymerase Dpo4 from Sulfolobus solfataricus. Although full-length product formation was observed, steady-state kinetic data show that dATP insertion opposite DFT is greatly inhibited relative to insertion opposite T (approximately 5,000-fold). No products were observed in the pre-steady-state. Furthermore, it is noteworthy that Dpo4 strongly prefers dATP opposite DFT over dGTP (approximately 200-fold) and that the polymerase is able to extend an A:DFT but not a G:DFT pair. We present crystal structures of Dpo4 in complex with DNA duplexes containing the DFT analog, the first for any DNA polymerase. In the structures, template-DFT is either positioned opposite primer-A or -G at the -1 site or is unopposed by a primer base and followed by a dGTP:A mismatch pair at the active site, representative of a -1 frameshift. The three structures provide insight into the discrimination by Dpo4 between dATP and dGTP opposite DFT and its inability to extend beyond a G:DFT pair. Although hydrogen bonding is clearly important for error-free replication by this Y-class DNA polymerase, our work demonstrates that Dpo4 also relies on shape and electrostatics to distinguish between correct and incorrect incoming nucleotide.
- Department of Biochemistry and Center in Molecular Toxicology, Vanderbilt University School of Medicine, Nashville, Tennessee 37232-0146, USA.
Organizational Affiliation: