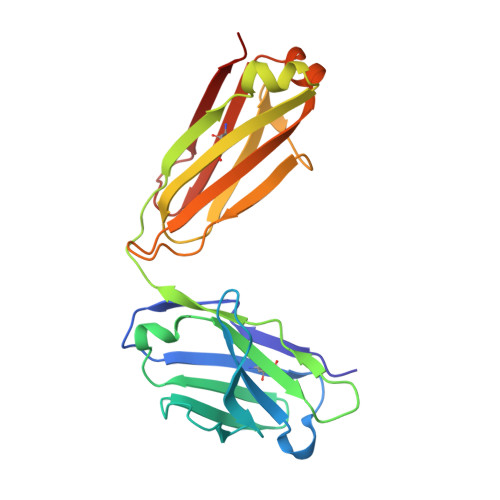
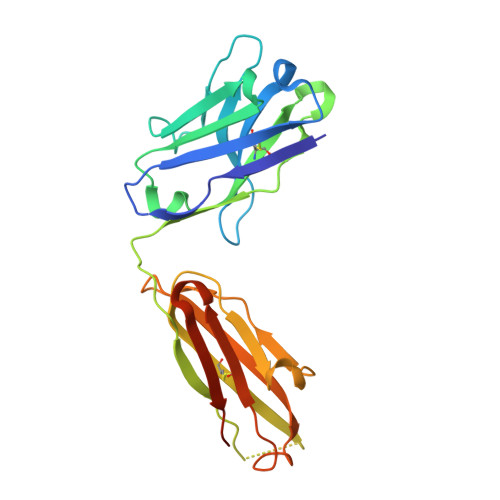
Structure of the Recombinant Antibody Fab Fragment F3P4.
Frey, D., Huber, T., Plueckthun, A., Gruetter, M.G.(2008) Acta Crystallogr D Biol Crystallogr 64: 636
- PubMed: 18560151
- DOI: https://doi.org/10.1107/S0907444908007282
- Primary Citation of Related Structures:
2V7N - PubMed Abstract:
The structure of the antibody Fab fragment f3p4, which was selected from a subset of the synthetic HuCAL antibody library to bind the sodium citrate symporter CitS, is described at 1.92 A resolution. Comparison with computational models revealed deviations in a few framework positions and in the binding loops. The crystals belong to space group P2(1)2(1)2 and contain four molecules in the asymmetric unit, with unit-cell parameters a=102.77, b=185.92, c=102.97 A. These particular unit-cell parameters allowed pseudo-merohedral twinning; interestingly, the twinning law relates a twofold screw axis to a twofold axis.
- Department of Biochemistry, University of Zürich, Switzerland.
Organizational Affiliation: