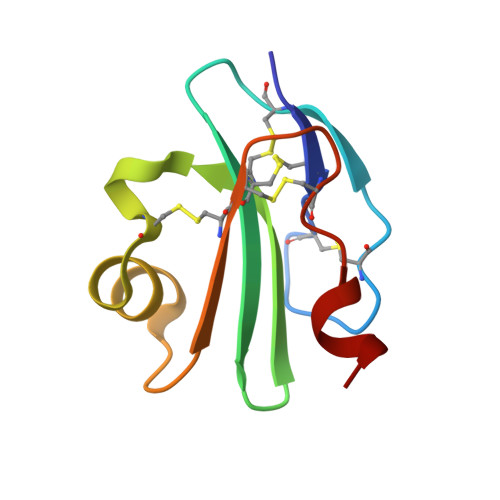
High-Resolution Structures of Bacterially Expressed Soluble Human Cd59.
Leath, K.J., Johnson, S., Roversi, P., Hughes, T.R., Smith, R.A.G., Mackenzie, L., Morgan, B.P., Lea, S.M.(2007) Acta Crystallogr Sect F Struct Biol Cryst Commun 63: 648
- PubMed: 17671359
- DOI: https://doi.org/10.1107/S1744309107033477
- Primary Citation of Related Structures:
2J8B, 2UWR, 2UX2 - PubMed Abstract:
CD59 is a membrane-bound glycoprotein that protects host cells from lysis by inhibiting the terminal pathway of complement, preventing the formation and insertion of the membrane attack complex (MAC). Crystals of bacterially expressed and nonglycosylated recombinant soluble human CD59 have been obtained from three crystallization conditions, each of which gave rise to a distinct crystal form. Each crystal form led to a crystal structure at high resolution (1.15, 1.35 and 1.8 A). In one of these structures the electron-density map shows an as yet unidentified small molecule in the predicted C8/C9-binding site. The presence/absence of this ligand is linked to alternate conformations of the amino acids implicated in C8/C9 binding.
- Sir William Dunn School of Pathology, University of Oxford, Oxford OX1 3RE, England.
Organizational Affiliation: