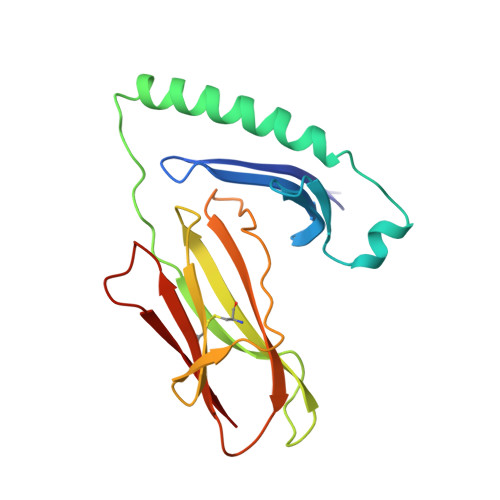
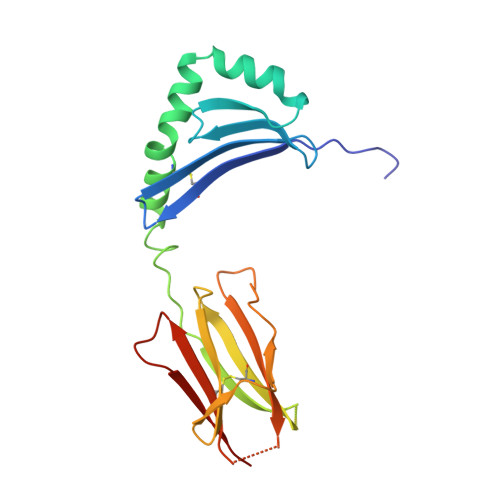
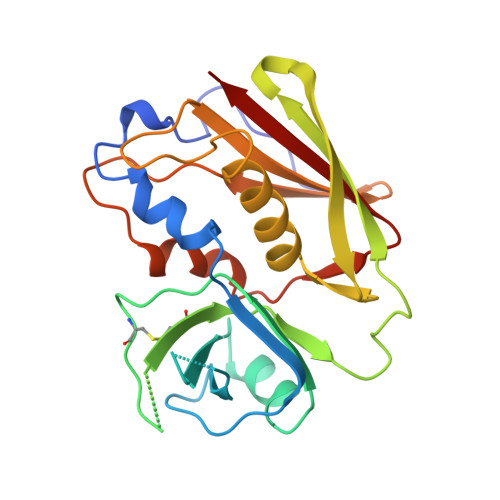
X-ray crystal structure of HLA-DR4 (DRA*0101, DRB1*0401) complexed with a peptide from human collagen II.
Dessen, A., Lawrence, C.M., Cupo, S., Zaller, D.M., Wiley, D.C.(1997) Immunity 7: 473-481
- PubMed: 9354468
- DOI: https://doi.org/10.1016/s1074-7613(00)80369-6
- Primary Citation of Related Structures:
2SEB - PubMed Abstract:
Genetic predisposition to rheumatoid arthritis (RA) is linked to the MHC class II allele HLA-DR4. The charge of the amino acid at DRbeta71 in the peptide-binding site appears to be critical in discriminating DR molecules linked to increased disease susceptibility. We have determined the 2.5 A x-ray structure of the DR4 molecule with the strongest linkage to RA (DRB1*0401) complexed with a human collagen II peptide. Details of a predicted salt bridge between lysine DRbeta71 and aspartic acid at the P4 peptide position suggest how it may participate in both antigen binding and TCR activation. A model is proposed for the DR4 recognition of collagen II (261-273), an antigen immunodominant in human-transgenic mouse models of RA.
- Laboratory of Molecular Medicine, The Children's Hospital, Boston, Massachusetts 02115, USA. dessen@xtal22.tch.harvard.edu
Organizational Affiliation: