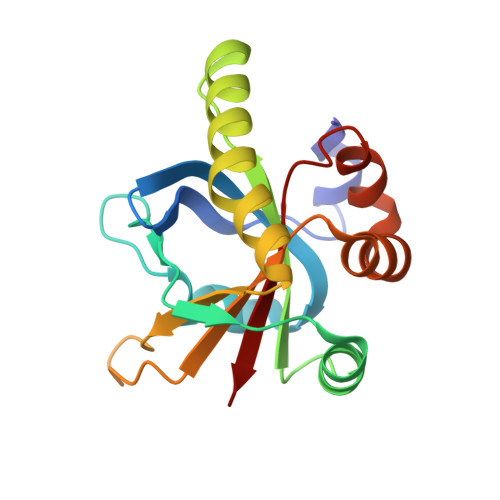
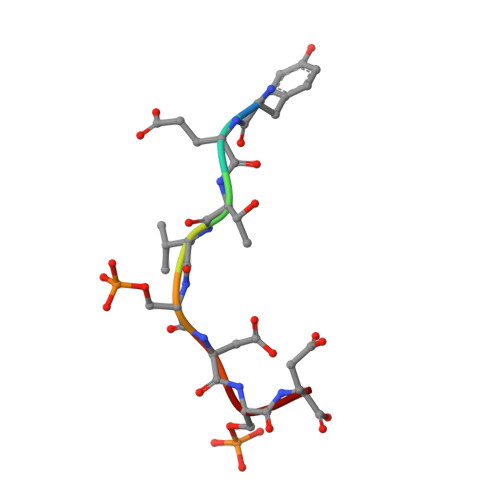
Structural insights into the recruitment of SMRT by the corepressor SHARP under phosphorylative regulation.
Mikami, S., Kanaba, T., Takizawa, N., Kobayashi, A., Maesaki, R., Fujiwara, T., Ito, Y., Mishima, M.(2014) Structure 22: 35-46
- PubMed: 24268649
- DOI: https://doi.org/10.1016/j.str.2013.10.007
- Primary Citation of Related Structures:
2RT5 - PubMed Abstract:
The transcriptional corepressors SMRT/NCoR, components of histone deacetylase complexes, interact with nuclear receptors and many other transcription factors. SMRT is a target for the ubiquitously expressed protein kinase CK2, which is known to phosphorylate a wide variety of substrates. Increasing evidence suggests that CK2 plays a regulatory role in many cellular events, particularly, in transcription. However, little is known about the precise mode of action involved. Here, we report the three-dimensional structure of a SMRT/HDAC1-associated repressor protein (SHARP) in complex with phosphorylated SMRT, as determined by solution NMR. Phosphorylation of the CK2 site on SMRT significantly increased affinity for SHARP. We also confirmed the significance of CK2 phosphorylation by reporter assay and propose a mechanism involving the process of phosphorylation acting as a molecular switch. Finally, we propose that the SPOC domain functions as a phosphorylation binding module.
- Graduate School of Science and Engineering, Tokyo Metropolitan University, 1-1 Minamiosawa Hachioji 192-0397, Japan.
Organizational Affiliation: