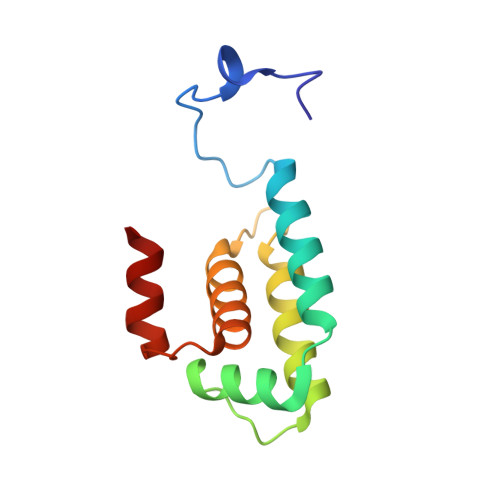
Solution structures and DNA binding properties of the N-terminal SAP domains of SUMO E3 ligases from Saccharomyces cerevisiae and Oryza sativa.
Suzuki, R., Shindo, H., Tase, A., Kikuchi, Y., Shimizu, M., Yamazaki, T.(2009) Proteins 75: 336-347
- PubMed: 18831036
- DOI: https://doi.org/10.1002/prot.22243
- Primary Citation of Related Structures:
2RNN, 2RNO - PubMed Abstract:
SUMO E3 ligase of the Siz/PIAS family that promotes sumoylation of target proteins contains SAP motif in its N-terminal region. The SAP motif with a consensus sequence of 35 residues was first proposed to be as a new DNA binding motif found in diverse nuclear proteins involved in chromosomal organization. We have determined solution structures of the SAP domains of SUMO ligases Siz1 from yeast and rice by NMR spectroscopy, showing that the structure of the SAP domain (residues 2-105) of rice Siz1 is a four-helix bundle with an up-down-extended loop-down-up topology, whereas the SAP domain (residues 1-111) of yeast Siz1 is comprised of five helices where the fifth helix alpha5 causes a significant change in the alignment of the four-helix bundle characteristic to the SAP domains of the Siz/PIAS family. We have also demonstrated that both SAP domains have binding ability to an A/T-rich DNA, but that binding affinity of yeast Siz1 SAP is at least by an order of magnitude higher than that of rice Siz1 SAP. Our NMR titration experiments clearly showed that yeast Siz1 SAP uses alpha2-helix for DNA binding more effectively than rice Siz1 SAP, which would result from the dislocation of this helix due to the existence of the extra helix alpha5. In addition, based on the structures of the SAP domains determined here and registered in Protein Data Bank, general features of structures of the SAP domains are discussed in conjunction with equivocal nature of their DNA binding.
- Protein Research Unit, National Institute of Agrobiological Sciences, Ibaraki 305-8602, Japan.
Organizational Affiliation: