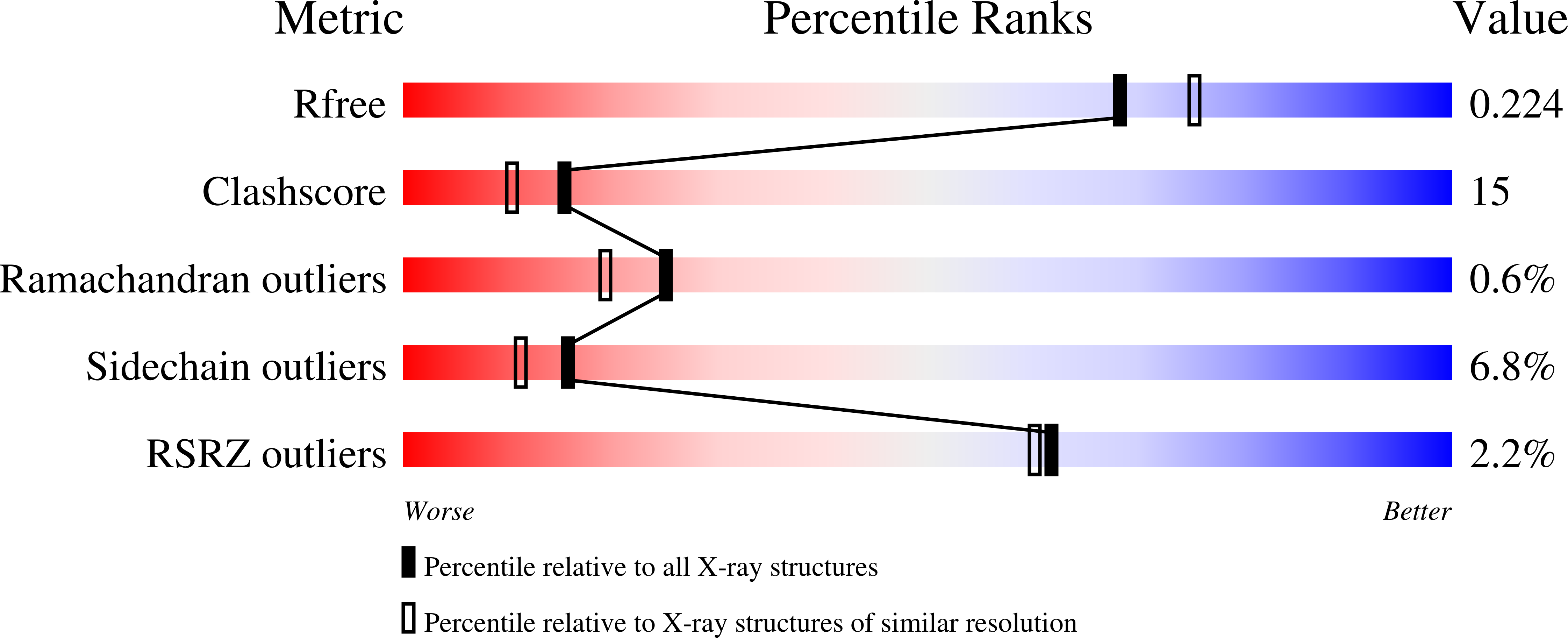
Crystal structures of murine thrombin in complex with the extracellular fragments of murine protease-activated receptors PAR3 and PAR4.
Bah, A., Chen, Z., Bush-Pelc, L.A., Mathews, F.S., Di Cera, E.(2007) Proc Natl Acad Sci U S A 104: 11603-11608
- PubMed: 17606903
- DOI: https://doi.org/10.1073/pnas.0704409104
- Primary Citation of Related Structures:
2PUX, 2PV9 - PubMed Abstract:
It has been proposed that the cleaved form of protease-activated receptor 3 (PAR3) acts as a cofactor for thrombin cleavage and activation of PAR4 on murine platelets, but the molecular basis of this physiologically important effect remains elusive. X-ray crystal structures of murine thrombin bound to extracellular fragments of the murine receptors PAR3 ((38)SFNGGPQNTFEEFPLSDIE(56)) and PAR4 ((51)KSSDKPNPR downward arrow GYPGKFCANDSDTLELPASSQA(81), downward arrow = site of cleavage) have been solved at 2.0 and 3.5 A resolution, respectively. The cleaved form of PAR3, traced in the electron density maps from Gln-44 to Glu-56, makes extensive hydrophobic and electrostatic contacts with exosite I of thrombin through the fragment (47)FEEFPLSDIE(56). Occupancy of exosite I by PAR3 allosterically changes the conformation of the 60-loop and shifts the position of Trp-60d approximately 10 A with a resulting widening of the access to the active site. The PAR4 fragment, traced entirely in the electron density maps except for five C-terminal residues, clamps Trp-60d, Tyr-60a, and the aryl-binding site of thrombin with Pro-56 and Pro-58 at the P2 and P4 positions and engages the primary specificity pocket with Arg-59. The fragment then leaves the active site with Gly-60 and folds into a short helical turn that directs the backbone away from exosite I and over the autolysis loop. The structures demonstrate that thrombin activation of PAR4 may occur with exosite I available to bind cofactor molecules, like the cleaved form of PAR3, whose function is to promote substrate diffusion into the active site by allosterically changing the conformation of the 60-loop.
- Department of Biochemistry and Molecular Biophysics, Washington University School of Medicine, Box 8231, St. Louis, MO 63110, USA.
Organizational Affiliation: