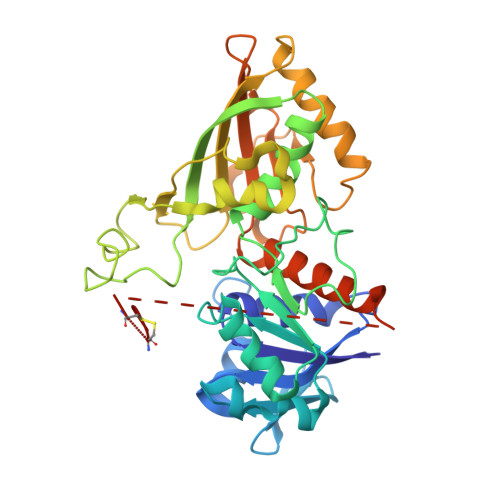
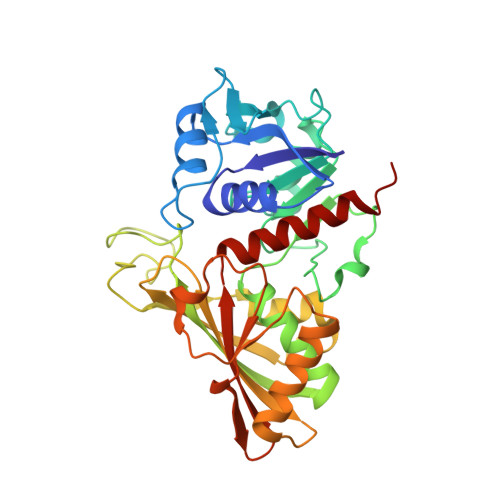
Molecular mechanism of thioredoxin regulation in photosynthetic A2B2-glyceraldehyde-3-phosphate dehydrogenase.
Fermani, S., Sparla, F., Falini, G., Martelli, P.L., Casadio, R., Pupillo, P., Ripamonti, A., Trost, P.(2007) Proc Natl Acad Sci U S A 104: 11109-11114
- PubMed: 17573533
- DOI: https://doi.org/10.1073/pnas.0611636104
- Primary Citation of Related Structures:
2PKQ, 2PKR - PubMed Abstract:
Chloroplast glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a light-regulated, NAD(P)H-dependent enzyme involved in plant photosynthetic carbon reduction. Unlike lower photosynthetic organisms, which only contain A(4)-GAPDH, the major GAPDH isoform of land plants is made up of A and B subunits, the latter containing a C-terminal extension (CTE) with fundamental regulatory functions. Light-activation of AB-GAPDH depends on the redox state of a pair of cysteines of the CTE, which can form a disulfide bond under control of thioredoxin f, leading to specific inhibition of the NADPH-dependent activity. The tridimensional structure of A(2)B(2)-GAPDH from spinach chloroplasts, crystallized in the oxidized state, shows that each disulfide-containing CTE is docked into a deep cleft between a pair of A and B subunits. The structure of the CTE was derived from crystallographic data and computational modeling and confirmed by site-specific mutagenesis. Structural analysis of oxidized A(2)B(2)-GAPDH and chimeric mutant [A+CTE](4)-GAPDH revealed that Arg-77, which is essential for coenzyme specificity and high NADPH-dependent activity, fails to interact with NADP in these kinetically inhibited GAPDH tetramers and is attracted instead by negative residues of oxidized CTE. Other subtle changes in catalytic domains and overall conformation of the tetramers were noticed in oxidized A(2)B(2)-GAPDH and [A+CTE](4)-GAPDH, compared with fully active A(4)-GAPDH. The CTE is envisioned as a redox-sensitive regulatory domain that can force AB-GAPDH into a kinetically inhibited conformation under oxidizing conditions, which also occur during dark inactivation of the enzyme in vivo.
- Department of Chemistry, University of Bologna, Via Selmi 2, 40126 Bologna, Italy.
Organizational Affiliation: