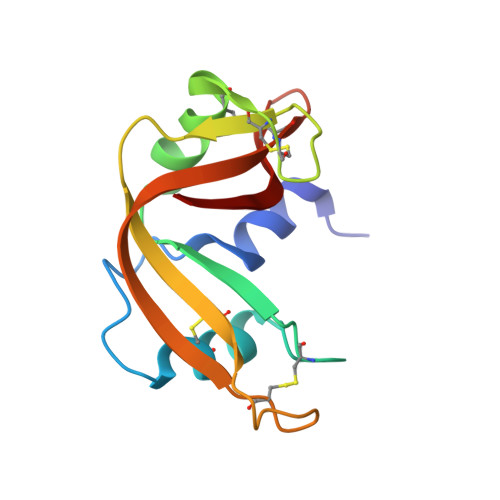
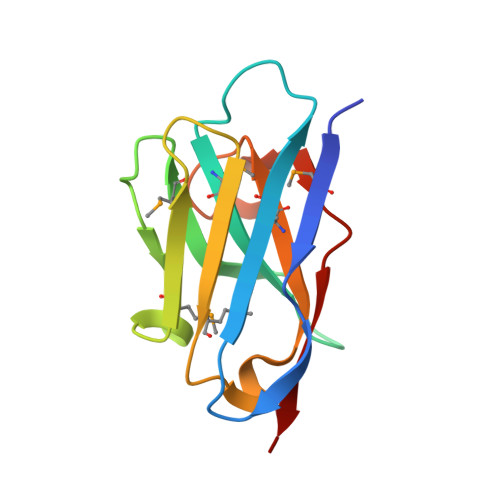
Toward chaperone-assisted crystallography: protein engineering enhancement of crystal packing and X-ray phasing capabilities of a camelid single-domain antibody (VHH) scaffold
Tereshko, V., Uysal, S., Koide, A., Margalef, K., Koide, S., Kossiakoff, A.A.(2008) Protein Sci 17: 1175-1187
- PubMed: 18445622
- DOI: https://doi.org/10.1110/ps.034892.108
- Primary Citation of Related Structures:
2P42, 2P43, 2P44, 2P45, 2P46, 2P47, 2P48 - PubMed Abstract:
A crystallization chaperone is an auxiliary protein that binds to a target of interest, enhances and modulates crystal packing, and provides high-quality phasing information. We critically evaluated the effectiveness of a camelid single-domain antibody (V(H)H) as a crystallization chaperone. By using a yeast surface display system for V(H)H, we successfully introduced additional Met residues in the core of the V(H)H scaffold. We identified a set of SeMet-labeled V(H)H variants that collectively produced six new crystal forms as the complex with the model antigen, RNase A. The crystals exhibited monoclinic, orthorhombic, triclinic, and tetragonal symmetry and have one or two complexes in the asymmetric unit, some of which diffracted to an atomic resolution. The phasing power of the Met-enriched V(H)H chaperone allowed for auto-building the entire complex using single-anomalous dispersion technique (SAD) without the need for introducing SeMet into the target protein. We show that phases produced by combining SAD and V(H)H model-based phases are accurate enough to easily solve structures of the size reported here, eliminating the need to collect multiple wavelength multiple-anomalous dispersion (MAD) data. Together with the presence of high-throughput selection systems (e.g., phage display libraries) for V(H)H, the enhanced V(H)H domain described here will be an excellent scaffold for producing effective crystallization chaperones.
- Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, Illinois 60637, USA.
Organizational Affiliation: