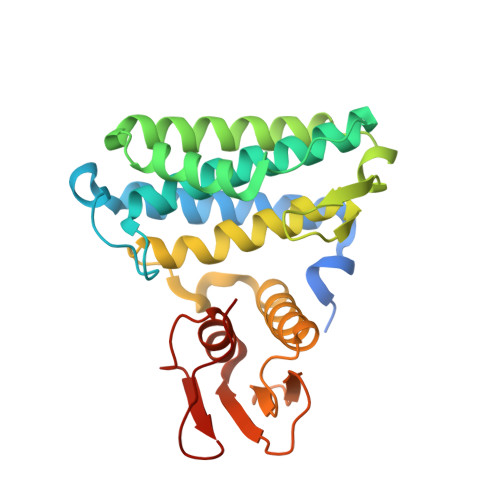
Crystal Structures and Site-directed Mutagenesis of a Mycothiol-dependent Enzyme Reveal a Novel Folding and Molecular Basis for Mycothiol-mediated Maleylpyruvate Isomerization
Wang, R., Yin, Y.J., Wang, F., Li, M., Feng, J., Zhang, H.M., Zhang, J.P., Liu, S.J., Chang, W.R.(2007) J Biological Chem 282: 16288-16294
- PubMed: 17428791
- DOI: https://doi.org/10.1074/jbc.M610347200
- Primary Citation of Related Structures:
2NSF, 2NSG - PubMed Abstract:
Mycothiol (MSH) is the major low molecular mass thiols in many Gram-positive bacteria such as Mycobacterium tuberculosis and Corynebacterium glutamicum. The physiological roles of MSH are believed to be equivalent to those of GSH in Gram-negative bacteria, but current knowledge of MSH is limited to detoxification of alkalating chemicals and protection from host cell defense/killing systems. Recently, an MSH-dependent maleylpyruvate isomerase (MDMPI) was discovered from C. glutamicum, and this isomerase represents one example of many putative MSH-dependent enzymes that take MSH as cofactor. In this report, fourteen mutants of MDMPI were generated. The wild type and mutant (H52A) MDMPIs were crystallized and their structures were solved at 1.75 and 2.05 A resolution, respectively. The crystal structures reveal that this enzyme contains a divalent metal-binding domain and a C-terminal domain possessing a novel folding pattern (alphabetaalphabetabetaalpha fold). The divalent metal-binding site is composed of residues His52, Glu144, and His148 and is located at the bottom of a surface pocket. Combining the structural and site-directed mutagenesis studies, it is proposed that this surface pocket including the metal ion and MSH moiety formed the putative catalytic center.
- National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Organizational Affiliation: