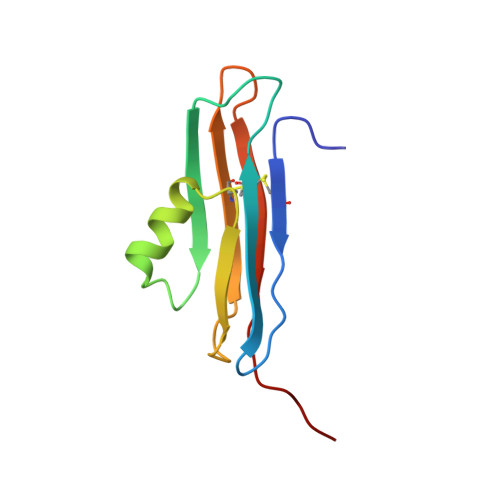
Solution structure of the coxsackievirus and adenovirus receptor domain 2
Jiang, S., Caffrey, M.(2007) Protein Sci 16: 539-542
- PubMed: 17322536
- DOI: https://doi.org/10.1110/ps.062643507
- Primary Citation of Related Structures:
2NPL - PubMed Abstract:
The coxsackievirus and adenovirus receptor (CAR) mediates entry of coxsackievirus and adenovirus. CAR possesses an extracellular region that is comprised of 2 immunoglobulin domains termed CAR-D1 and CAR-D2. In the present work, the solution structure of CAR-D2, consisting of residues 142-235 of human CAR, has been determined by NMR spectroscopy. CAR-D2 is shown to be a beta-sandwich motif comprised of two beta-sheets, which are stabilized by two disulfide bonds. The first beta-sheet is comprised of beta-strands A, B, and E, and the second beta-sheet is comprised of beta-strands C, F, and G. A relatively hydrophobic helix is found between beta-strands C and E, which replaces beta-strand D of the classical c-type immunoglobulin fold.
- Department of Biochemistry and Molecular Genetics, University of Illinois at Chicago, Chicago, Illinois 60607, USA.
Organizational Affiliation: