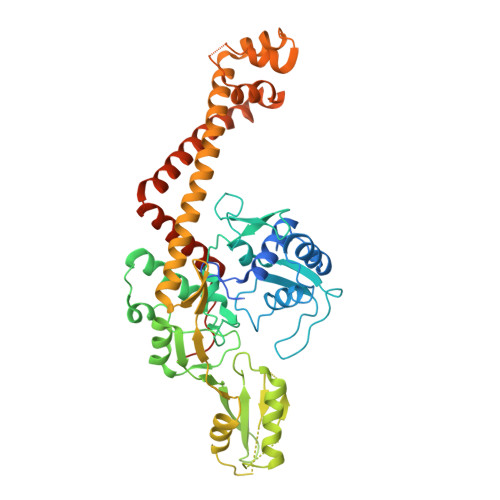
Breakage-Reunion Domain of Streptococcus pneumoniae Topoisomerase IV: Crystal Structure of a Gram-Positive Quinolone Target.
Laponogov, I., Veselkov, D.A., Sohi, M.K., Pan, X.S., Achari, A., Yang, C., Ferrara, J.D., Fisher, L.M., Sanderson, M.R.(2007) PLoS One 2: e301-e301
- PubMed: 17375187
- DOI: https://doi.org/10.1371/journal.pone.0000301
- Primary Citation of Related Structures:
2NOV - PubMed Abstract:
The 2.7 A crystal structure of the 55-kDa N-terminal breakage-reunion domain of topoisomerase (topo) IV subunit A (ParC) from Streptococcus pneumoniae, the first for the quinolone targets from a gram-positive bacterium, has been solved and reveals a 'closed' dimer similar in fold to Escherichia coli DNA gyrase subunit A (GyrA), but distinct from the 'open' gate structure of Escherichia coli ParC. Unlike GyrA whose DNA binding groove is largely positively charged, the DNA binding site of ParC exhibits a distinct pattern of alternating positively and negatively charged regions coincident with the predicted positions of the grooves and phosphate backbone of DNA. Based on the ParC structure, a new induced-fit model for sequence-specific recognition of the gate (G) segment by ParC has been proposed. These features may account for the unique DNA recognition and quinolone targeting properties of pneumococcal type II topoisomerases compared to their gram-negative counterparts.
- Randall Division of Cell and Molecular Biophysics, School of Biomedical and Health Sciences, King's College London, London, United Kindgom.
Organizational Affiliation: