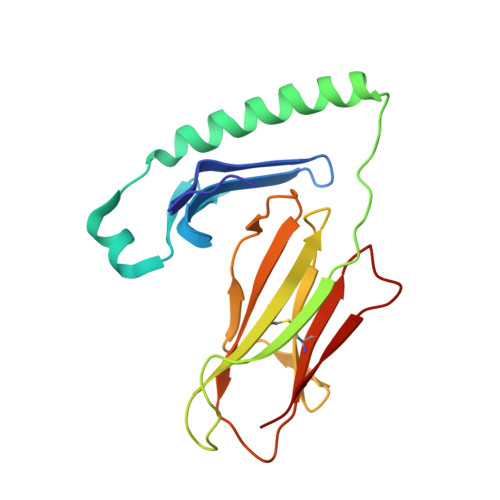
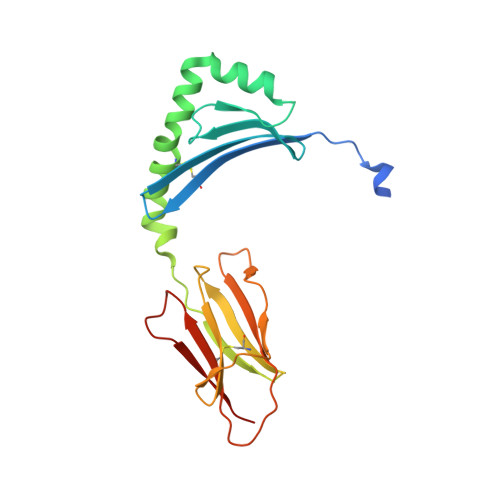
A structural and immunological basis for the role of human leukocyte antigen DQ8 in celiac disease
Henderson, K.N., Tye-Din, J.A., Reid, H.H., Chen, Z., Borg, N.A., Beissbarth, T., Tatham, A., Mannering, S.I., Purcell, A.W., Dudek, N.L., van Heel, D.A., McCluskey, J., Rossjohn, J., Anderson, R.P.(2007) Immunity 27: 23-34
- PubMed: 17629515
- DOI: https://doi.org/10.1016/j.immuni.2007.05.015
- Primary Citation of Related Structures:
2NNA - PubMed Abstract:
The risk of celiac disease is strongly associated with human leukocyte antigen (HLA) DQ2 and to a lesser extent with HLA DQ8. Although the pathogenesis of HLA-DQ2-mediated celiac disease is established, the underlying basis for HLA-DQ8-mediated celiac disease remains unclear. We showed that T helper 1 (Th1) responses in HLA-DQ8-associated celiac pathology were indeed HLA DQ8 restricted and that multiple, mostly deamidated peptides derived from protease-sensitive sites of gliadin were recognized. This pattern of reactivity contrasted with the more absolute deamidation dependence and relative protease resistance of the dominant gliadin peptide in DQ2-mediated disease. We provided a structural basis for the selection of HLA-DQ8-restricted, deamidated gliadin peptides. The data established that the molecular mechanisms underlying HLA-DQ8-mediated celiac disease differed markedly from the HLA-DQ2-mediated form of the disease. Accordingly, nondietary therapeutic interventions in celiac disease might need to be tailored to the genotype of the individual.
- The Protein Crystallography Unit, Department of Biochemistry and Molecular Biology, School of Biomedical Sciences, Monash University, Clayton, Victoria 3800, Australia.
Organizational Affiliation: