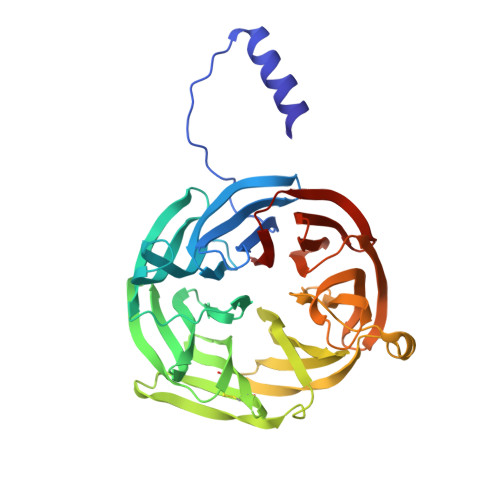
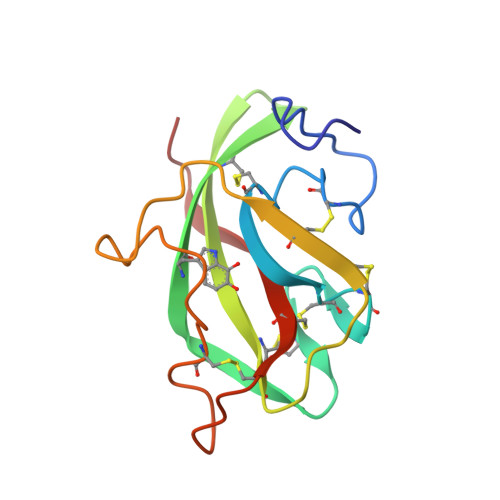
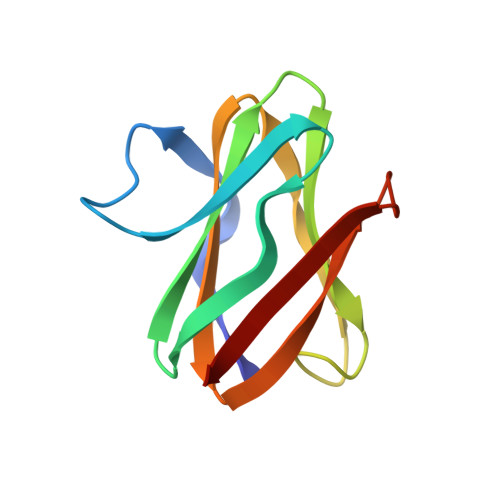
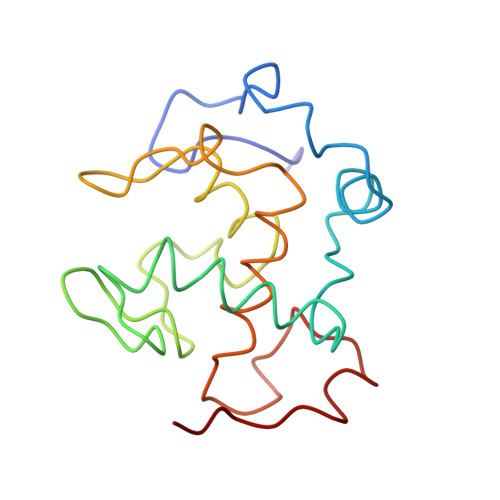
Structure of an electron transfer complex: methylamine dehydrogenase, amicyanin, and cytochrome c551i.
Chen, L., Durley, R.C., Mathews, F.S., Davidson, V.L.(1994) Science 264: 86-90
- PubMed: 8140419
- DOI: https://doi.org/10.1126/science.8140419
- Primary Citation of Related Structures:
2MTA - PubMed Abstract:
The crystal structure of a ternary protein complex has been determined at 2.4 angstrom resolution. The complex is composed of three electron transfer proteins from Paracoccus denitrificans, the quinoprotein methylamine dehydrogenase, the blue copper protein amicyanin, and the cytochrome c551i. The central region of the c551i is folded similarly to several small bacterial c-type cytochromes; there is a 45-residue extension at the amino terminus and a 25-residue extension at the carboxyl terminus. The methylamine dehydrogenase-amicyanin interface is largely hydrophobic, whereas the amicyanin-cytochrome interface is more polar, with several charged groups present on each surface. Analysis of the simplest electron transfer pathways between the redox partners points out the importance of other factors such as energetics in determining the electron transfer rates.
- Department of Biochemistry and Molecular Biophysics, Washington University School of Medicine, St. Louis, MO 63110.
Organizational Affiliation: