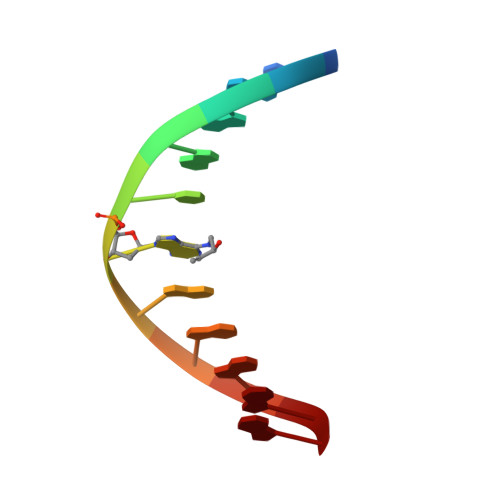
Major Groove Orientation of the (2S)-N(6)-(2-Hydroxy-3-buten-1-yl)-2'-deoxyadenosine DNA Adduct Induced by 1,2-Epoxy-3-butene.
Kowal, E.A., Wickramaratne, S., Kotapati, S., Turo, M., Tretyakova, N., Stone, M.P.(2014) Chem Res Toxicol 27: 1675-1686
- PubMed: 25238403
- DOI: https://doi.org/10.1021/tx500159w
- Primary Citation of Related Structures:
2MNX - PubMed Abstract:
1,3-Butadiene (BD) is an environmental and occupational toxicant classified as a human carcinogen. It is oxidized by cytochrome P450 monooxygenases to 1,2-epoxy-3-butene (EB), which alkylates DNA. BD exposures lead to large numbers of mutations at A:T base pairs even though alkylation of guanines is more prevalent, suggesting that one or more adenine adducts of BD play a role in BD-mediated genotoxicity. However, the etiology of BD-mediated genotoxicity at adenine remains poorly understood. EB alkylates the N(6) exocyclic nitrogen of adenine to form N(6)-(hydroxy-3-buten-1-yl)-2'-dA ((2S)-N(6)-HB-dA) adducts ( Tretyakova , N. , Lin , Y. , Sangaiah , R. , Upton , P. B. , and Swenberg , J. A. ( 1997 ) Carcinogenesis 18 , 137 - 147 ). The structure of the (2S)-N(6)-HB-dA adduct has been determined in the 5'-d(C(1)G(2)G(3)A(4)C(5)Y(6)A(7)G(8)A(9)A(10)G(11))-3':5'-d(C(12)T(13)T(14)C(15)T(16)T(17)G(18)T(19) C(20)C(21)G(22))-3' duplex [Y = (2S)-N(6)-HB-dA] containing codon 61 (underlined) of the human N-ras protooncogene, from NMR spectroscopy. The (2S)-N(6)-HB-dA adduct was positioned in the major groove, such that the butadiene moiety was oriented in the 3' direction. At the Cα carbon, the methylene protons of the modified nucleobase Y(6) faced the 5' direction, which placed the Cβ carbon in the 3' direction. The Cβ hydroxyl group faced toward the solvent, as did carbons Cγ and Cδ. The Cβ hydroxyl group did not form hydrogen bonds with either T(16) O(4) or T(17) O(4). The (2S)-N(6)-HB-dA nucleoside maintained the anti conformation about the glycosyl bond, and the modified base retained Watson-Crick base pairing with the complementary base (T(17)). The adduct perturbed stacking interactions at base pairs C(5):G(18), Y(6):T(17), and A(7):T(16) such that the Y(6) base did not stack with its 5' neighbor C(5), but it did with its 3' neighbor A(7). The complementary thymine T(17) stacked well with both 5' and 3' neighbors T(16) and G(18). The presence of the (2S)-N(6)-HB-dA resulted in a 5 °C reduction in the Tm of the duplex, which is attributed to less favorable stacking interactions and adduct accommodation in the major groove.
- Department of Chemistry, Center in Molecular Toxicology, Vanderbilt Ingram Cancer Center, and Center for Structural Biology, Vanderbilt University , 2201 West End Avenue, Nashville, Tennessee 37235, United States.
Organizational Affiliation: