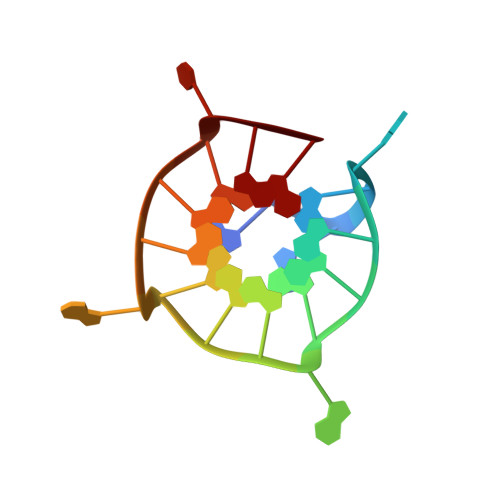
Structure and Conformational Dynamics of a Stacked Dimeric G-Quadruplex Formed by the Human CEB1 Minisatellite.
Adrian, M., Ang, D.J., Lech, C.J., Heddi, B., Nicolas, A., Phan, A.T.(2014) J Am Chem Soc 136: 6297-6305
- PubMed: 24742225
- DOI: https://doi.org/10.1021/ja4125274
- Primary Citation of Related Structures:
2MB4 - PubMed Abstract:
CEB1 is a highly polymorphic human minisatellite. In yeast, the size variation of CEB1 tandem arrays has been associated with the capacity of the motif to form G-quadruplexes. Here we report on the NMR solution structure of a G-quadruplex formed by the CEB1 DNA G-rich fragment d(AGGGGGGAGGGAGGGTGG), harboring several G-tracts including one with six continuous guanines. This sequence forms a dimeric G-quadruplex involving the stacking of two subunits, each being a unique snapback parallel-stranded scaffold with three G-tetrad layers, three double-chain-reversal loops, and a V-shaped loop. The two subunits are stacked at their 5'-end tetrads, and multiple stacking rotamers may be present due to a high symmetry at the stacking interface. There is a conformational exchange in the millisecond time scale involving a swapping motion between two bases of the six-guanine tract. Our results not only add to the understanding of how the G-quadruplex formation in human minisatellite leads to genetic instability but also address the fundamental questions regarding stacking of G-quadruplexes and how a long continuous G-tract participates in the structure and conformational dynamics of G-quadruplexes.
- School of Physical and Mathematical Sciences, Nanyang Technological University , Singapore 637371, Singapore.
Organizational Affiliation: