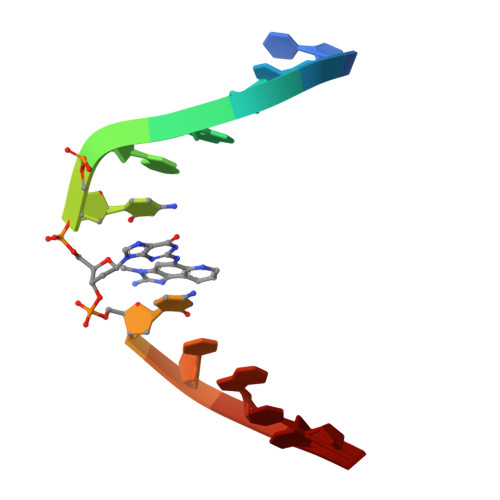
Base-displaced intercalation of the 2-amino-3-methylimidazo[4,5-f]quinolone N2-dG adduct in the NarI DNA recognition sequence.
Stavros, K.M., Hawkins, E.K., Rizzo, C.J., Stone, M.P.(2014) Nucleic Acids Res 42: 3450-3463
- PubMed: 24366876
- DOI: https://doi.org/10.1093/nar/gkt1109
- Primary Citation of Related Structures:
2MAV - PubMed Abstract:
2-Amino-3-methylimidazo[4,5-f]quinolone (IQ), a heterocyclic amine found in cooked meats, undergoes bioactivation to a nitrenium ion, which alkylates guanines at both the C8-dG and N2-dG positions. The conformation of a site-specific N2-dG-IQ adduct in an oligodeoxynucleotide duplex containing the iterated CG repeat restriction site of the NarI endonuclease has been determined. The IQ moiety intercalates, with the IQ H4a and CH3 protons facing the minor groove, and the IQ H7a, H8a and H9a protons facing the major groove. The adducted dG maintains the anti-conformation about the glycosyl bond. The complementary dC is extruded into the major groove. The duplex maintains its thermal stability, which is attributed to stacking between the IQ moiety and the 5'- and 3'-neighboring base pairs. This conformation is compared to that of the C8-dG-IQ adduct in the same sequence, which also formed a 'base-displaced intercalated' conformation. However, the C8-dG-IQ adopted the syn conformation placing the Watson-Crick edge of the modified dG into the major groove. In addition, the C8-dG-IQ adduct was oriented with the IQ CH3 group and H4a and H5a facing the major groove. These differences may lead to differential processing during DNA repair and replication.
- Department of Chemistry, Center in Molecular Toxicology, Vanderbilt-Ingram Cancer Center, Vanderbilt Institute of Chemical Biology, Vanderbilt University, Nashville, TN 37235-1822, USA.
Organizational Affiliation: