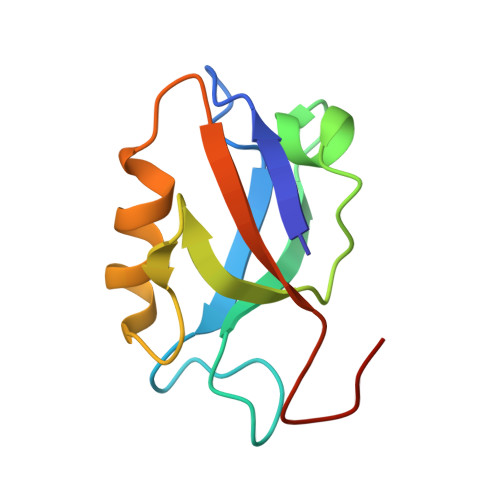
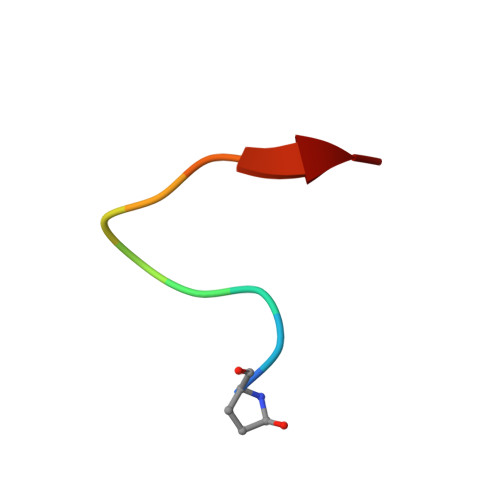
Structural insights into a wildtype domain of the oncoprotein E6 and its interaction with a PDZ domain.
Mischo, A., Ohlenschlager, O., Hortschansky, P., Ramachandran, R., Gorlach, M.(2013) PLoS One 8: e62584-e62584
- PubMed: 23638119
- DOI: https://doi.org/10.1371/journal.pone.0062584
- Primary Citation of Related Structures:
2M3L, 2M3M - PubMed Abstract:
The high-risk human papilloma virus (HPV) oncoproteins E6 and E7 interact with key cellular regulators and are etiological agents for tumorigenesis and tumor maintenance in cervical cancer and other malignant conditions. E6 induces degradation of the tumor suppressor p53, activates telomerase and deregulates cell polarity. Analysis of E6 derived from a number of high risk HPV finally yielded the first structure of a wild-type HPV E6 domain (PDB 2M3L) representing the second zinc-binding domain of HPV 51 E6 (termed 51Z2) determined by NMR spectroscopy. The 51Z2 structure provides clues about HPV-type specific structural differences between E6 proteins. The observed temperature sensitivity of the well-folded wild-type E6 domain implies a significant malleability of the oncoprotein in vivo. Hence, the structural differences between individual E6 and their malleability appear, together with HPV type-specific surface exposed side-chains, to provide the structural basis for the different interaction networks reported for individual E6 proteins. Furthermore, the interaction of 51Z2 with a PDZ domain of hDlg was analyzed. Human Dlg constitutes a prototypic representative of the large family of PDZ proteins regulating cell polarity, which are common targets of high-risk HPV E6. Nine C-terminal residues of 51Z2 interact with the second PDZ domain of hDlg2. Surface plasmon resonance in conjunction with the NMR spectroscopy derived complex structure (PDB 2M3M) indicate that E6 residues N-terminal to the canonical PDZ-BM of E6 significantly contribute to this interaction and increase affinity. The structure of the complex reveals how residues outside of the classical PDZ-BM enhance the affinity of E6 towards PDZ domains. Such mechanism facilitates successful competition of E6 with cellular PDZ-binding proteins and may apply to PDZ-binding proteins of other viruses as well.
- Biomolecular NMR Spectroscopy, Leibniz Institute for Age Research - Fritz Lipmann Institute, Jena, Germany.
Organizational Affiliation: