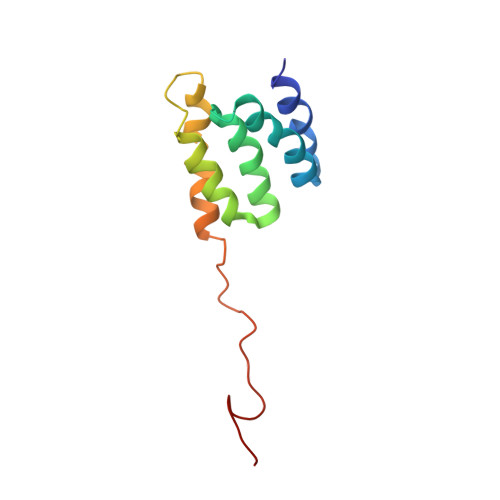
High-Resolution Structural Analysis Shows How Tah1 Tethers Hsp90 to the R2TP Complex.
Back, R., Dominguez, C., Rothe, B., Bobo, C., Beaufils, C., Morera, S., Meyer, P., Charpentier, B., Branlant, C., Allain, F.H., Manival, X.(2013) Structure 21: 1834-1847
- PubMed: 24012479
- DOI: https://doi.org/10.1016/j.str.2013.07.024
- Primary Citation of Related Structures:
2LSU, 2LSV - PubMed Abstract:
The ubiquitous Hsp90 chaperone participates in snoRNP and RNA polymerase assembly through interaction with the R2TP complex. This complex includes the proteins Tah1, Pih1, Rvb1, and Rvb2. Tah1 bridges Hsp90 to R2TP. Its minimal TPR domain includes two TPR motifs and a capping helix. We established the high-resolution solution structures of Tah1 free and in complex with the Hsp90 C-terminal peptide. The TPR fold is similar in the free and bound forms and we show experimentally that in addition to its solvating/stabilizing role, the capping helix is essential for the recognition of the Hsp90 (704)EMEEVD(709) motif. In addition to Lys79 and Arg83 from the carboxylate clamp, this helix bears Tyr82 forming a π/S-CH3 interaction with Hsp90 M(705) from the peptide 310 helix. The Tah1 C-terminal region is unfolded, and we demonstrate that it is essential for the recruitment of the Pih1 C-terminal domain and folds upon binding.
- Ingénierie Moléculaire et Physiopathologie Articulaire (IMoPA), UMR 7365 Université de Lorraine-CNRS, Biopôle de l'Université de Lorraine, Campus Biologie Santé, 9 Avenue de la forêt de Haye, BP 184, 54505 Vandœuvre-lès-Nancy, France.
Organizational Affiliation: