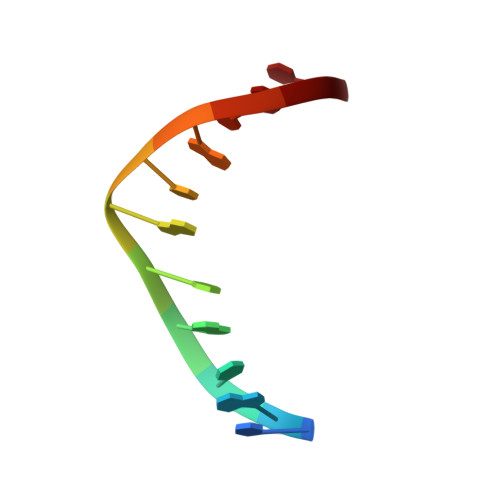
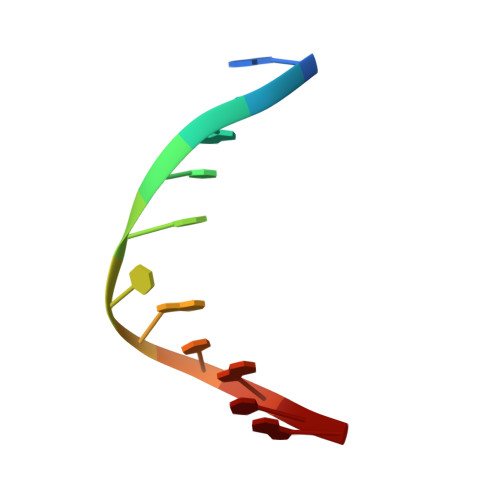
Structure and stability of DNA containing an aristolactam II-dA lesion: implications for the NER recognition of bulky adducts.
Lukin, M., Zaliznyak, T., Johnson, F., de Los Santos, C.(2012) Nucleic Acids Res 40: 2759-2770
- PubMed: 22121223
- DOI: https://doi.org/10.1093/nar/gkr1094
- Primary Citation of Related Structures:
2LGM - PubMed Abstract:
Aristolochic acids I and II are prevalent plant toxicants found in the Aristolochiaceae plant family. Metabolic activation of the aristolochic acids leads to the formation of a cyclic N-hydroxylactam product that can react with the peripheral amino group of purine bases generating bulky DNA adducts. These lesions are mutagenic and established human carcinogens. Interestingly, although AL-dG adducts progressively disappear from the DNA of laboratory animals, AL-dA lesions has lasting persistence in the genome. We describe here NMR structural studies of an undecameric duplex damaged at its center by the presence of an ALII-dA adduct. Our data establish a locally perturbed double helical structure that accommodates the bulky adduct by displacing the counter residue into the major groove and stacking the ALII moiety between flanking bases. The presence of the ALII-dA perturbs the conformation of the 5'-side flanking base pair, but all other pairs of the duplex adopt standard conformations. Thermodynamic studies reveal that the lesion slightly decreases the energy of duplex formation in a sequence-dependent manner. We discuss our results in terms of its implications for the repair of ALII-dA adducts in mammalian cells.
- Department of Pharmacological Sciences, School of Medicine, Stony Brook University, Stony Brook, NY 11794-8651, USA.
Organizational Affiliation: