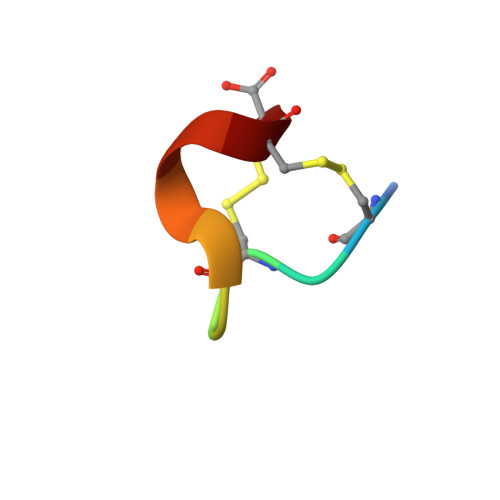
Pc16a, the first characterized peptide from Conus pictus venom, shows a novel disulfide connectivity.
Van Der Haegen, A., Peigneur, S., Dyubankova, N., Moller, C., Mari, F., Diego-Garcia, E., Naude, R., Lescrinier, E., Herdewijn, P., Tytgat, J.(2012) Peptides 34: 106-113
- PubMed: 22079220
- DOI: https://doi.org/10.1016/j.peptides.2011.10.026
- Primary Citation of Related Structures:
2LER - PubMed Abstract:
A novel conotoxin, pc16a, was isolated from the venom of Conus pictus. This is the first peptide characterized from this South-African cone snail and it has only 11 amino acid residues, SCSCKRNFLCC*, with the rare cysteine framework XVI and a monoisotopic mass of 1257.6Da. Two peptides were synthesized with two possible conformations: globular (pc16a_1) and ribbon (pc16a_2). pc16a_1 co-eluted with the native peptide, which indicates a disulfide connectivity I-III, II-IV. The structure of pc16a_1 was determined by NMR. Both synthetic peptides were used to elucidate the biological activity. Bioassays were performed on crickets, ghost shrimps, larvae of the mealworm beetle and mice, but no effect was seen. Using two-electrode voltage clamp, a range of voltage-gated ion channels (Na(v) and K(v)) and nicotinic acetylcholine receptors were screened, but again no activity was found. Hence, the specific target of pc16a still remains to be discovered.
- University of Leuven, Herestraat 49, Leuven, Belgium.
Organizational Affiliation: