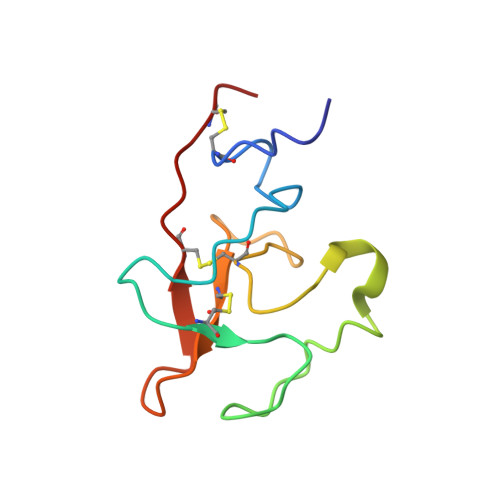
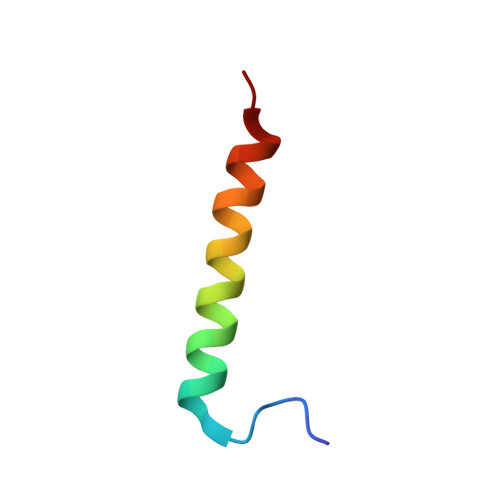
Solution structure of the complex of VEK-30 and plasminogen kringle 2.
Wang, M., Zajicek, J., Geiger, J.H., Prorok, M., Castellino, F.J.(2010) J Struct Biol 169: 349-359
- PubMed: 19800007
- DOI: https://doi.org/10.1016/j.jsb.2009.09.011
- Primary Citation of Related Structures:
2KJ4 - PubMed Abstract:
The solution structure of the complex containing the isolated kringle 2 domain of human plasminogen (K2(Pg)) and VEK-30, a 30-amino acid residue internal peptide from a streptococcal M-like plasminogen (Pg) binding protein (PAM), has been determined by multinuclear high-resolution NMR. Complete backbone and side-chain assignments were obtained from triple-resonance experiments, after which structure calculations were performed and ultimately refined by restrained molecular simulation in water. We find that, in contrast with the dimer of complexes observed in the asymmetric unit of the crystal, global correlation times and buoyant molecular weight determinations of the complex and its individual components showed the monomeric nature of all species in solution. The NMR-derived structure of K2(Pg) in complex with VEK-30 presents a folding pattern typical of other kringle domains, while bound VEK-30 forms an end-to-end alpha-helix (residues 6-27) in the complex. Most of the VEK-30/K2(Pg) interactions in solution occur between a single face of the alpha-helix of VEK-30 and the lysine binding site (LBS) of K2(Pg). The canonical LBS of K2(Pg), consisting of Asp54, Asp56, Trp60, Arg69, and Trp70 (kringle numbering), interacts with an internal pseudo-lysine of VEK-30, comprising side-chains of Arg17, His18, and Glu20. Site-specific mutagenesis analysis confirmed that the electrostatic field formed by the N-terminal anionic residues of the VEK-30 alpha-helix, viz., Asp7, and the non-conserved cationic residues of K2(Pg), viz., Lys43 and Arg55, play additional important roles in the docking of VEK-30 to K2(Pg). Structural analysis and kringle sequence alignments revealed several important features related to exosite binding that provide a structural rationale for the high specificity and affinity of VEK-30 for K2(Pg).
- W.M. Keck Center for Transgene Research, 230 Raclin-Carmichael Hall, University of Notre Dame, Notre Dame, IN 46556, USA.
Organizational Affiliation: