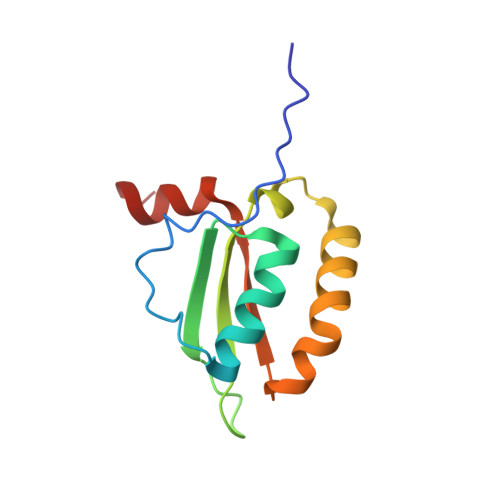
Solution-state NMR structure of the putative morphogene protein BolA (PFE0790c) from Plasmodium falciparum.
Buchko, G.W., Yee, A., Semesi, A., Myler, P.J., Arrowsmith, C.H., Hui, R.(2015) Acta Crystallogr Sect F Struct Biol Cryst Commun 71: 514-521
- PubMed: 25945703
- DOI: https://doi.org/10.1107/S2053230X1402799X
- Primary Citation of Related Structures:
2KDN - PubMed Abstract:
Protozoa of the genus Plasmodium are responsible for malaria, which is perhaps the most important parasitic disease to infect mankind. The emergence of Plasmodium strains resistant to current therapeutics and prophylactics makes the development of new treatment strategies urgent. Among the potential targets for new antimalarial drugs is the BolA-like protein PFE0790c from Plasmodium falciparum (Pf-BolA). While the function of BolA is unknown, it has been linked to cell morphology by regulating transcription in response to stress. Using an NMR-based method, an ensemble of 20 structures of Pf-BolA was determined and deposited in the PDB (PDB entry 2kdn). The overall topology of the Pf-BolA structure, α1-β1-β2-η1-α2/η2-β3-α3, with the β-strands forming a mixed β-sheet, is similar to the fold observed in other BolA structures. A helix-turn-helix motif similar to the class II KH fold associated with nucleic acid-binding proteins is present, but contains an FXGXXXL signature sequence that differs from the GXXG signature sequence present in class II KH folds, suggesting that the BolA family of proteins may use a novel protein-nucleic acid interface. A well conserved arginine residue, Arg50, hypothesized to play a role in governing the formation of the C-terminal α-helix in the BolA family of proteins, is too distant to form polar contacts with any side chains in this α-helix in Pf-BolA, suggesting that this conserved arginine may only serve a role in guiding the orientation of this C-terminal helix in some BolA proteins. A survey of BolA structures suggests that the C-terminal helix may not have a functional role and that the third helix (α2/η2) has a `kink' that appears to be conserved among the BolA protein structures. Circular dichroism spectroscopy shows that Pf-BolA is fairly robust, partially unfolding when heated to 353 K and refolding upon cooling to 298 K.
- Seattle Structural Genomics Center for Infectious Disease, USA.
Organizational Affiliation: