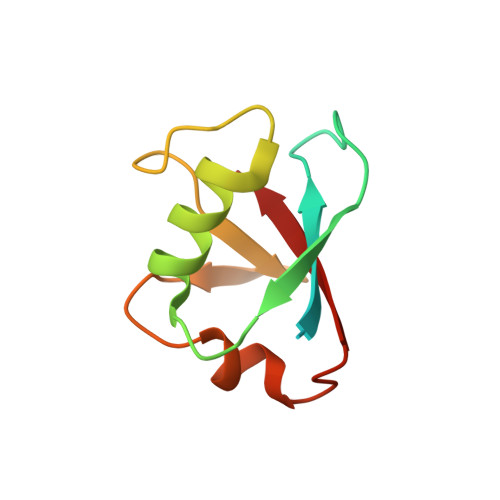
Structure of human homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 2 (Herpud2 or Herp)
Doherty, R., Wu, B., Fares, C., Lemak, A., Gutmanas, A., Semesi, A., Yee, A., Arrowsmith, C.H., Dhe-Paganon, S.To be published.