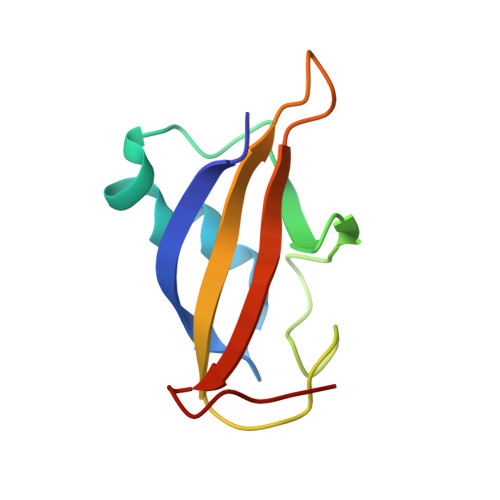
Identification and structural characterization of a CBP/p300-binding domain from the ETS family transcription factor GABP alpha
Kang, H.S., Nelson, M.L., Mackereth, C.D., Scharpf, M., Graves, B.J., McIntosh, L.P.(2008) J Mol Biology 377: 636-646
- PubMed: 18295234
- DOI: https://doi.org/10.1016/j.jmb.2008.01.054
- Primary Citation of Related Structures:
2JUO - PubMed Abstract:
Using NMR spectroscopy, we identified and characterized a previously unrecognized structured domain near the N-terminus (residues 35-121) of the ETS family transcription factor GABP alpha. The monomeric domain folds as a five-stranded beta-sheet crossed by a distorted helix. Although globally resembling ubiquitin, the GABP alpha fragment differs in its secondary structure topology and thus appears to represent a new protein fold that we term the OST (On-SighT) domain. The surface of the GABP alpha OST domain contains two predominant clusters of negatively-charged residues suggestive of electrostatically driven interactions with positively-charged partner proteins. Following a best-candidate approach to identify such a partner, we demonstrated through NMR-monitored titrations and glutathione S-transferase pulldown assays that the OST domain binds to the CH1 and CH3 domains of the co-activator histone acetyltransferase CBP/p300. This provides a direct structural link between GABP and a central component of the transcriptional machinery.
- Department of Biochemistry and Molecular Biology, University of British Columbia, Vancouver, BC, Canada.
Organizational Affiliation: