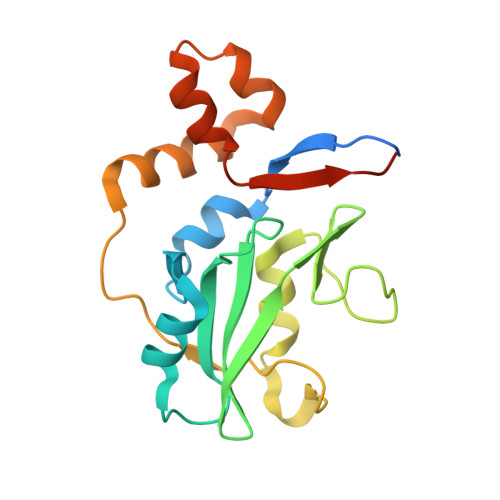
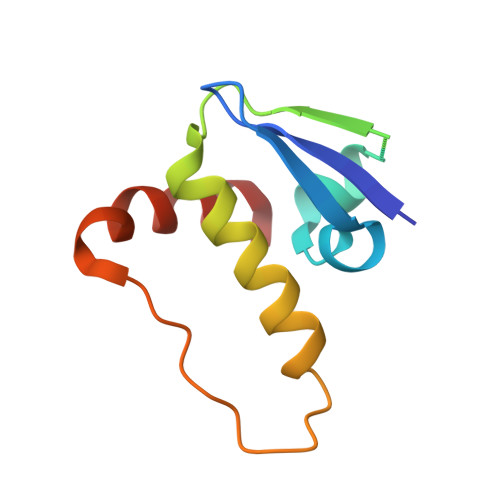
Structure of the SOCS4-ElonginB/C complex reveals a distinct SOCS box interface and the molecular basis for SOCS-dependent EGFR degradation.
Bullock, A.N., Rodriguez, M.C., Debreczeni, J.E., Songyang, Z., Knapp, S.(2007) Structure 15: 1493-1504
- PubMed: 17997974
- DOI: https://doi.org/10.1016/j.str.2007.09.016
- Primary Citation of Related Structures:
2IZV - PubMed Abstract:
Tyrosine kinase signaling is tightly controlled by negative feedback inhibitors including suppressors of cytokine signaling (SOCS). SOCS assemble as SH2 domain substrate recognition modules in ElonginB/C-cullin ubiquitin ligases. In accordance, SOCS4 reduces STAT3 signaling from EGFR through increased receptor degradation. Variable C-termini in SOCS4-SOCS7 exclude these family members from a SOCS2-type domain arrangement in which a strictly conserved C terminus determines domain packing. The structure of the SOCS4-ElonginC-ElonginB complex reveals a distinct SOCS structural class. The N-terminal ESS helix functionally replaces the CIS/SOCS1-SOCS3 family C terminus in a distinct SH2-SOCS box interface that facilitates further interdomain packing between the extended N- and C-terminal regions characteristic for this subfamily. Using peptide arrays and calorimetry the STAT3 site in EGFR (pY(1092)) was identified as a high affinity SOCS4 substrate (K(D) = 0.5 microM) revealing a mechanism for EGFR degradation. SOCS4 also bound JAK2 and KIT with low micromolar affinity, whereas SOCS2 was specific for GH-receptor.
- University of Oxford, Structural Genomics Consortium, Botnar Research Centre, Oxford OX3 7LD, United Kingdom. alex.bullock@sgc.ox.ac.uk
Organizational Affiliation: