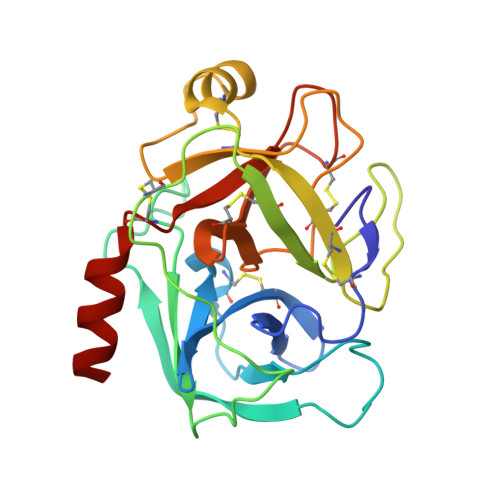
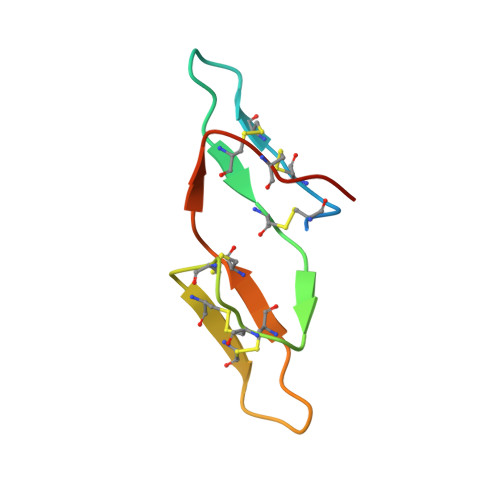
Crystal structure of the anticarcinogenic Bowman-Birk inhibitor from snail medic (Medicago scutellata) seeds complexed with bovine trypsin
Capaldi, S., Perduca, M., Faggion, B., Carrizo, M.E., Tava, A., Ragona, L., Monaco, H.L.(2007) J Struct Biol 158: 71-79
- PubMed: 17142058
- DOI: https://doi.org/10.1016/j.jsb.2006.10.017
- Primary Citation of Related Structures:
2ILN - PubMed Abstract:
The structure of the ternary complex of the anticarcinogenic Bowman-Birk protease inhibitor purified from snail medic (Medicago scutellata) seeds (MSTI) and two molecules of bovine trypsin has been solved by X-ray diffraction analysis of single crystals to a resolution of 2.0 A. This is the highest resolution model of a ternary complex of this type currently available. The two binding loops of the MSTI differ in only one amino acid and have in both cases an arginine in position P1. The distances between the residues of the inhibitor at the binding interface and the trypsin side chains that recognize them are almost identical in the two sites. When compared to the NMR model of the uncomplexed MSTI, the inhibitor in the functional assembly with trypsin shows the largest differences in the two P2' residues. Compared with the similar ternary complex of the soybean trypsin inhibitor, this model shows very small differences in the polypeptide chain of the trypsin binding sites and its largest difference in the area between Asp 26 and His 32 of the MSTI which in the soybean inhibitor has an extra Leu inserted in position 29.
- Biocrystallography Laboratory, Department of Science and Technology, University of Verona, Strada Le Grazie 15, 37134 Verona, Italy.
Organizational Affiliation: