Irditoxin, a novel covalently linked heterodimeric three-finger toxin with high taxon-specific neurotoxicity.
Pawlak, J., Mackessy, S.P., Sixberry, N.M., Stura, E.A., Le Du, M.H., Menez, R., Foo, C.S., Menez, A., Nirthanan, S., Kini, R.M.(2009) FASEB J 23: 534-545
- PubMed: 18952712
- DOI: https://doi.org/10.1096/fj.08-113555
- Primary Citation of Related Structures:
2H7Z - PubMed Abstract:
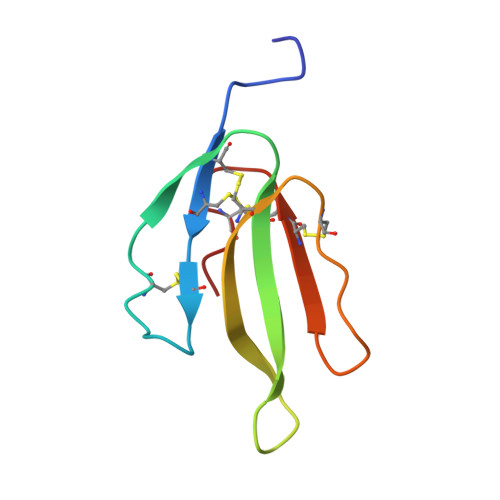
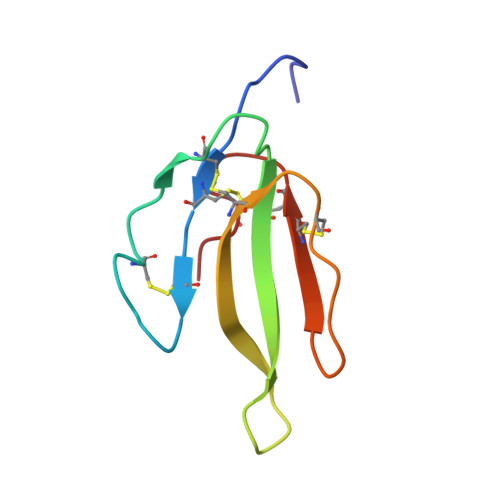
A novel heterodimeric three-finger neurotoxin, irditoxin, was isolated from venom of the brown treesnake Boiga irregularis (Colubridae). Irditoxin subunit amino acid sequences were determined by Edman degradation and cDNA sequencing. The crystal structure revealed two subunits with a three-finger protein fold, typical for "nonconventional" toxins such as denmotoxin, bucandin, and candoxin. This is the first colubrid three-finger toxin dimer, covalently connected via an interchain disulfide bond. Irditoxin showed taxon-specific lethality toward birds and lizards and was nontoxic toward mice. It produced a potent neuromuscular blockade at the avian neuromuscular junction (IC(50)=10 nM), comparable to alpha-bungarotoxin, but was three orders of magnitude less effective at the mammalian neuromuscular junction. Covalently linked heterodimeric three-finger toxins found in colubrid venoms constitute a new class of venom peptides, which may be a useful source of new neurobiology probes and therapeutic leads.
- Department of Biological Sciences, Faculty of Science, National University of Singapore, Science Dr. 4, Singapore 117543.
Organizational Affiliation: