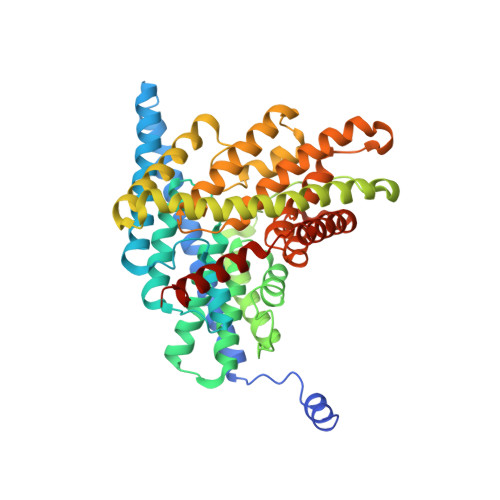
Uncoupling of a CLC Cl(-)/H(+) Exchange Transporter by Polyatomic Anions
Nguitragool, W., Miller, C.(2006) J Mol Biology 362: 682-690
- PubMed: 16905147
- DOI: https://doi.org/10.1016/j.jmb.2006.07.006
- Primary Citation of Related Structures:
2H2P, 2H2S - PubMed Abstract:
CLC-ec1 is a bacterial archetype of CLC transporters, a ubiquitous class of proteins that catalyze transmembrane exchange of Cl- and H+ necessary for pH regulation of numerous physiological processes. Despite a profusion of high-resolution structures, the molecular mechanism of exchange remains unknown. Here, we rigorously demonstrate strict exchange stoichiometry of 2 Cl-/1 H+. In addition to Cl- and Br-, two non-halide ions, NO3- and SCN-, are shown to be transported by CLC-ec1, but with reduced H+ counter-transport. The loss of proton coupling to these anions is accompanied by an absence of bound anions in the central and external Cl- binding sites in the protein's anion selectivity region, as revealed by crystallographic comparison of Br- and SeCN- bound to this region.
- Department of Biochemistry, Howard Hughes Medical Institute, Brandeis University, Waltham, MA 02454, USA.
Organizational Affiliation: