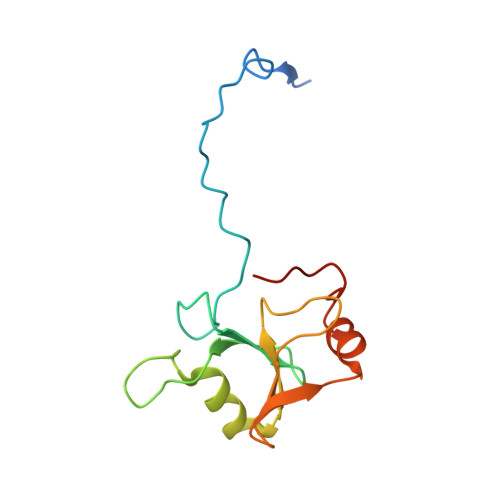
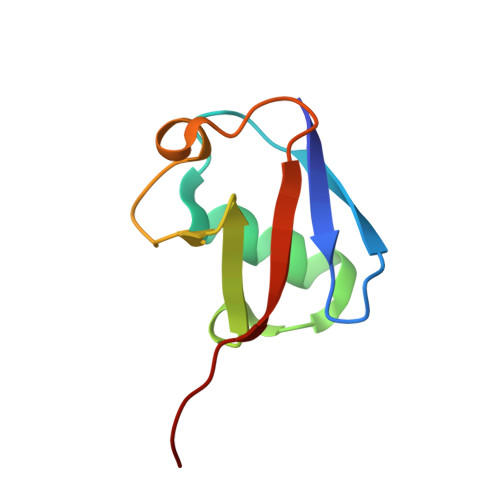
The Ubiquitin Binding Domain ZnF UBP Recognizes the C-Terminal Diglycine Motif of Unanchored Ubiquitin.
Reyes-Turcu, F.E., Horton, J.R., Mullally, J.E., Heroux, A., Cheng, X., Wilkinson, K.D.(2006) Cell 124: 1197-1208
- PubMed: 16564012
- DOI: https://doi.org/10.1016/j.cell.2006.02.038
- Primary Citation of Related Structures:
2G43, 2G45 - PubMed Abstract:
Ubiquitin binding proteins regulate the stability, function, and/or localization of ubiquitinated proteins. Here we report the crystal structures of the zinc-finger ubiquitin binding domain (ZnF UBP) from the deubiquitinating enzyme isopeptidase T (IsoT, or USP5) alone and in complex with ubiquitin. Unlike other ubiquitin binding domains, this domain contains a deep binding pocket where the C-terminal diglycine motif of ubiquitin is inserted, thus explaining the specificity of IsoT for an unmodified C terminus on the proximal subunit of polyubiquitin. Mutations in the domain demonstrate that it is required for optimal catalytic activation of IsoT. This domain is present in several other protein families, and the ZnF UBP domain from an E3 ligase also requires the C terminus of ubiquitin for binding. These data suggest that binding the ubiquitin C terminus may be necessary for the function of other proteins.
- Department of Biochemistry, Emory University School of Medicine, Atlanta, GA 30322, USA.
Organizational Affiliation: