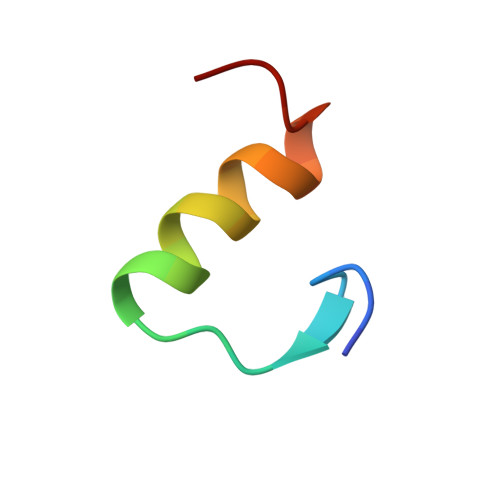
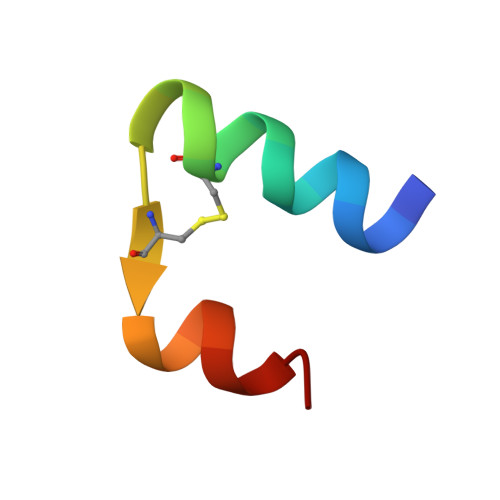
Solution structure and novel insights into the determinants of the receptor specificity of human relaxin-3.
Rosengren, K.J., Lin, F., Bathgate, R.A., Tregear, G.W., Daly, N.L., Wade, J.D., Craik, D.J.(2006) J Biological Chem 281: 5845-5851
- PubMed: 16365033
- DOI: https://doi.org/10.1074/jbc.M511210200
- Primary Citation of Related Structures:
2FHW - PubMed Abstract:
Relaxin-3 is the most recently discovered member of the relaxin family of peptide hormones. In contrast to relaxin-1 and -2, whose main functions are associated with pregnancy, relaxin-3 is involved in neuropeptide signaling in the brain. Here, we report the solution structure of human relaxin-3, the first structure of a relaxin family member to be solved by NMR methods. Overall, relaxin-3 adopts an insulin-like fold, but the structure differs crucially from the crystal structure of human relaxin-2 near the B-chain terminus. In particular, the B-chain C terminus folds back, allowing Trp(B27) to interact with the hydrophobic core. This interaction partly blocks the conserved RXXXRXXI motif identified as a determinant for the interaction with the relaxin receptor LGR7 and may account for the lower affinity of relaxin-3 relative to relaxin for this receptor. This structural feature is likely important for the activation of its endogenous receptor, GPCR135.
- Institute for Molecular Bioscience, University of Queensland, Brisbane, Queensland 4072, Australia.
Organizational Affiliation: