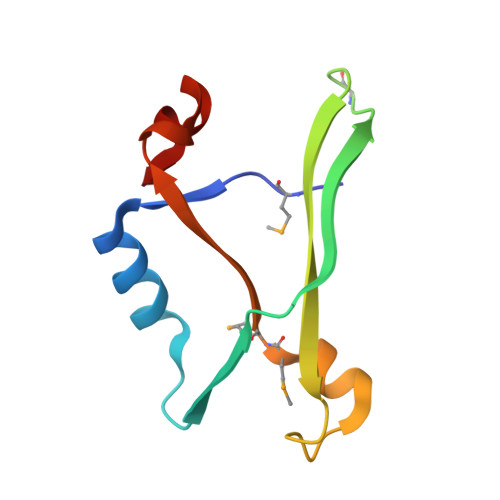
The crystal structure of the AF2331 protein from Archaeoglobus fulgidus DSM 4304 forms an unusual interdigitated dimer with a new type of alpha + beta fold.
Wang, S., Kirillova, O., Chruszcz, M., Gront, D., Zimmerman, M.D., Cymborowski, M.T., Shumilin, I.A., Skarina, T., Gorodichtchenskaia, E., Savchenko, A., Edwards, A.M., Minor, W.(2009) Protein Sci 18: 2410-2419
- PubMed: 19768810
- DOI: https://doi.org/10.1002/pro.251
- Primary Citation of Related Structures:
2FDO - PubMed Abstract:
The structure of AF2331, a 11-kDa orphan protein of unknown function from Archaeoglobus fulgidus, was solved by Se-Met MAD to 2.4 A resolution. The structure consists of an alpha + beta fold formed by an unusual homodimer, where the two core beta-sheets are interdigitated, containing strands alternating from both subunits. The decrease in solvent-accessible surface area upon dimerization is unusually large (3960 A(2)) for a protein of its size. The percentage of the total surface area buried in the interface (41.1%) is one of the largest observed in a nonredundant set of homodimers in the PDB and is above the mean for nearly all other types of homo-oligomers. AF2331 has no sequence homologs, and no structure similar to AF2331 could be found in the PDB using the CE, TM-align, DALI, or SSM packages. The protein has been identified in Pfam 23.0 as the archetype of a new superfamily and is topologically dissimilar to all other proteins with the "3-Layer (BBA) Sandwich" fold in CATH. Therefore, we propose that AF2331 forms a novel alpha + beta fold. AF2331 contains multiple negatively charged surface clusters and is located on the same operon as the basic protein AF2330. We hypothesize that AF2331 and AF2330 may form a charge-stabilized complex in vivo, though the role of the negatively charged surface clusters is not clear.
- Department of Molecular Physiology and Biological Physics, University of Virginia, Charlottesville, Virginia 22908, USA.
Organizational Affiliation: