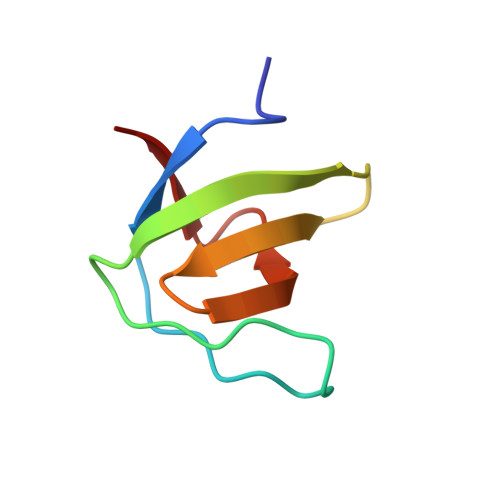
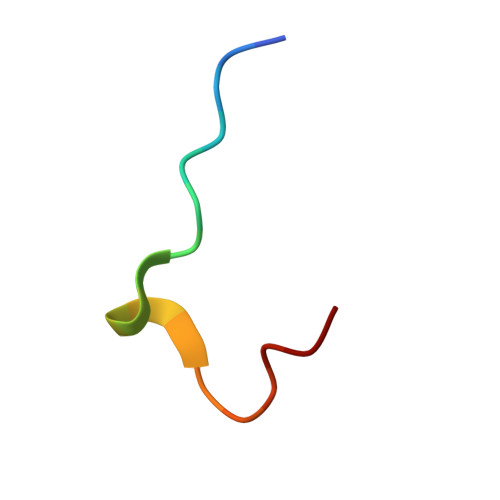
Crystal Structure of the SH3 Domain of betaPIX in Complex with a High Affinity Peptide from PAK2
Hoelz, A., Janz, J.M., Lawrie, S.D., Corwin, B., Lee, A., Sakmar, T.P.(2006) J Mol Biology 358: 509-522
- PubMed: 16527308
- DOI: https://doi.org/10.1016/j.jmb.2006.02.027
- Primary Citation of Related Structures:
2DF6, 2G6F - PubMed Abstract:
The p21-activated kinases (PAKs) are important effector proteins of the small GTPases Cdc42 and Rac and control cytoskeletal rearrangements and cell proliferation. The direct interaction of PAKs with guanine nucleotide exchange factors from the PIX/Cool family, which is responsible for the localization of PAK kinases to focal complexes in the cell, is mediated by a 24-residue peptide segment in PAKs and an N-terminal src homology 3 (SH3) domain in PIX/Cool. The SH3-binding segment of PAK contains the atypical consensus-binding motif PxxxPR, which is required for unusually high affinity binding. In order to understand the structural basis for the high affinity and specificity of the PIX-PAK interaction, we solved crystal structures for the N-terminal SH3 domain of betaPIX and for the complex of the atypical binding segment of PAK2 with the N-terminal SH3 domain of betaPIX at 0.92 A and 1.3A resolution, respectively. The asymmetric unit of the crystal contains two SH3 domains and two peptide ligands. The bound peptide adopts a conformation that allows for intimate contacts with three grooves on the surface of the SH3 domain that lie between the n-Src and RT-loops. Most notably, the arginine residue of the PxxxPR motif forms a salt-bridge and is tightly coordinated by a number of residues in the SH3 domain. This arginine-specific interaction appears to be the key determinant for the high affinity binding of PAK peptides. Furthermore, C-terminal residues of the peptide engage in additional interactions with the surface of the RT-loop, which significantly increases binding specificity. Compared to a recent NMR structure of a similar complex, our crystal structure reveals an alternate binding mode. Finally, we compare our crystal structure with the recently published betaPIX/Cbl-b complex structure, and suggest the existence of a molecular switch.
- The Rockefeller University, 1230 York Avenue, New York, NY 10021, USA. hoelza@rockefeller.edu
Organizational Affiliation: