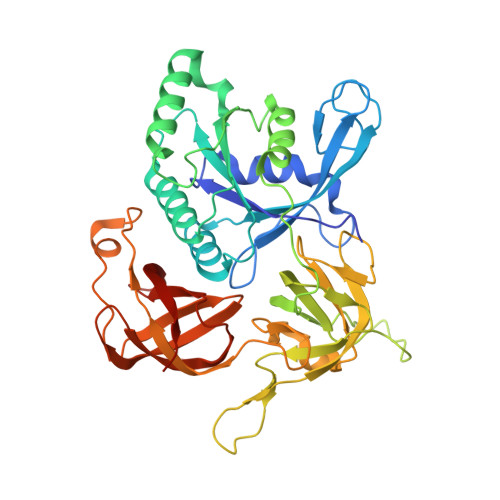
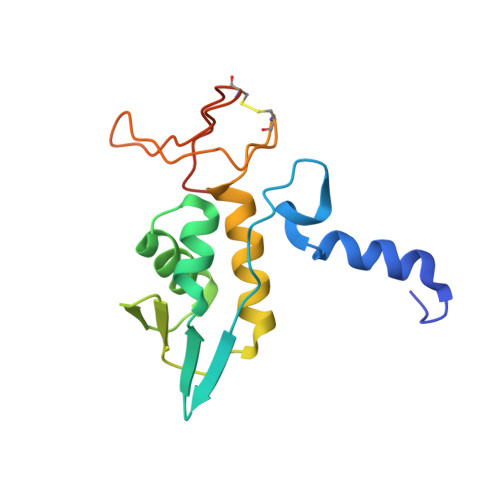
Structure of archaeal translational initiation factor 2 betagamma-GDP reveals significant conformational change of the beta-subunit and switch 1 region.
Sokabe, M., Yao, M., Sakai, N., Toya, S., Tanaka, I.(2006) Proc Natl Acad Sci U S A 103: 13016-13021
- PubMed: 16924118
- DOI: https://doi.org/10.1073/pnas.0604165103
- Primary Citation of Related Structures:
2D74, 2DCU - PubMed Abstract:
Archaeal/eukaryotic initiation factor 2 (a/eIF2) consists of alpha-, beta-, and gamma-subunits and delivers initiator methionine tRNA (Met-tRNA(i)) to a small ribosomal subunit in a GTP-dependent manner. The structures of the aIF2betagamma (archaeal initiation factor 2 betagamma) heterodimeric complex in the apo and GDP forms were analyzed at 2.8- and 3.4-A resolution, respectively. The results showed that the N-terminal helix and the central helix-turn-helix domain of the beta-subunit bind to the G domain of the gamma-subunit but are distant from domains 2 and 3, to which the alpha-subunit and Met-tRNA(i) bind. This result is consistent with most of the previous analyses of eukaryotic factors, and thus indicates that the binding mode is essentially conserved among a/eIF2. Comparison with the uncomplexed structure showed significant differences between the two forms of the beta-subunit, particularly the C-terminal zinc-binding domain, which does not interact with the gamma-subunit and was suggested previously to be involved in GTP hydrolysis. Furthermore, the switch 1 region in the gamma-subunit, which is shown to be responsible for Met-tRNA(i) binding by mutational analysis, is moved away from the nucleotide through the interaction with highly conserved R87 in the beta-subunit. These results implicate that conformational change of the beta-subunit facilitates GTP hydrolysis by inducing the conformational change of the switch 1 region toward the off state.
- Faculty of Advanced Life Sciences, Hokkaido University, Sapporo 060-0810, Japan.
Organizational Affiliation: