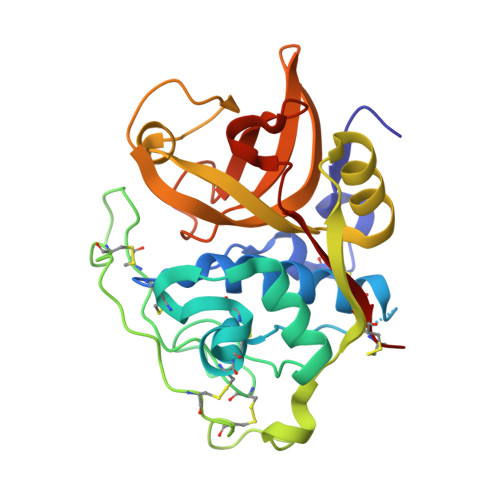
Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
Watanabe, D., Yamamoto, A., Matsumoto, K., Murata, M., Kitamura, K., Tomoo, K., Ishida, T.To be published.