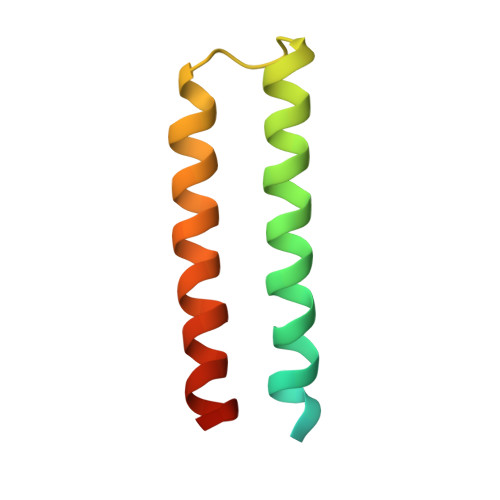
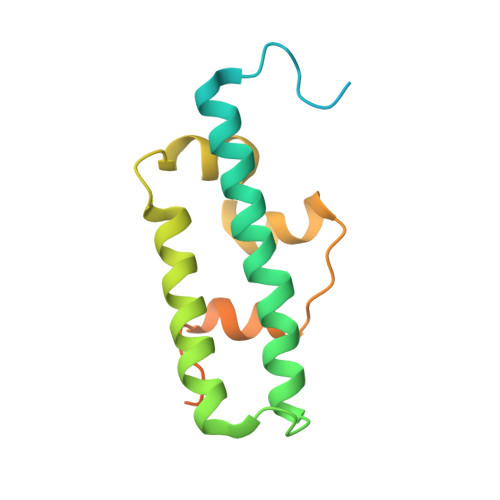
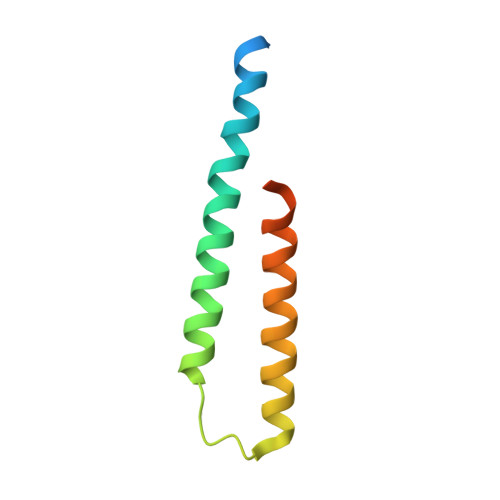
Escrt-I Core and Escrt-II Glue Domain Structures Reveal Role for Glue in Linking to Escrt-I and Membranes.
Teo, H., Gill, D.J., Sun, J., Perisic, O., Veprintsev, D.B., Wallis, Y., Emr, S.D., Williams, R.L.(2006) Cell 125: 99
- PubMed: 16615893
- DOI: https://doi.org/10.1016/j.cell.2006.01.047
- Primary Citation of Related Structures:
2CAY, 2CAZ - PubMed Abstract:
ESCRT complexes form the main machinery driving protein sorting from endosomes to lysosomes. Currently, the picture regarding assembly of ESCRTs on endosomes is incomplete. The structure of the conserved heterotrimeric ESCRT-I core presented here shows a fan-like arrangement of three helical hairpins, each corresponding to a different subunit. Vps23/Tsg101 is the central hairpin sandwiched between the other subunits, explaining the critical role of its "steadiness box" in the stability of ESCRT-I. We show that yeast ESCRT-I links directly to ESCRT-II, through a tight interaction of Vps28 (ESCRT-I) with the yeast-specific zinc-finger insertion within the GLUE domain of Vps36 (ESCRT-II). The crystal structure of the GLUE domain missing this insertion reveals it is a split PH domain, with a noncanonical lipid binding pocket that binds PtdIns3P. The simultaneous and reinforcing interactions of ESCRT-II GLUE domain with membranes, ESCRT-I, and ubiquitin are critical for ubiquitinated cargo progression from early to late endosomes.
- MRC Laboratory of Molecular Biology, Medical Research Council Centre, Cambridge, CB2 2QH, United Kingdom. hlt@mrc-lmb.cam.ac.uk
Organizational Affiliation: