Application of Fragment Screening and Fragment Linking to the Discovery of Novel Thrombin Inhibitors
Howard, N., Abell, C., Blakemore, W., Chessari, G., Congreve, M., Howard, S., Jhoti, H., Murray, C.W., Seavers, L.C.A., Van Montfort, R.L.M.(2006) J Med Chem 49: 1346
- PubMed: 16480269
- DOI: https://doi.org/10.1021/jm050850v
- Primary Citation of Related Structures:
2C8W, 2C8X, 2C8Y, 2C8Z, 2C90, 2C93 - PubMed Abstract:
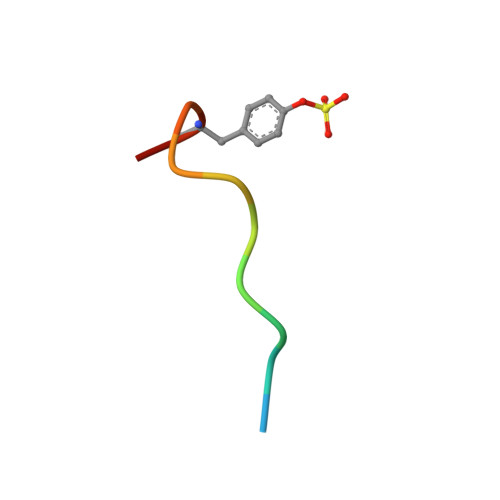
The screening of fragments is an alternative approach to high-throughput screening for the identification of leads for therapeutic targets. Fragment hits have been discovered using X-ray crystallographic screening of protein crystals of the serine protease enzyme thrombin. The fragment library was designed to avoid any well-precedented, strongly basic functionality. Screening hits included a novel ligand (3), which binds exclusively to the S2-S4 pocket, in addition to smaller fragments which bind to the S1 pocket. The structure of these protein-ligand complexes are presented. A chemistry strategy to link two such fragments together and to synthesize larger drug-sized compounds resulted in the efficient identification of hybrid inhibitors with nanomolar potency (e.g., 7, IC50 = 3.7 nM). These potent ligands occupy the same area of the active site as previously described peptidic inhibitors, while having very different chemical architecture.
- Astex Therapeutics, 436 Cambridge Science Park, Milton Road, Cambridge CB4 0QA, United Kingdom.
Organizational Affiliation: