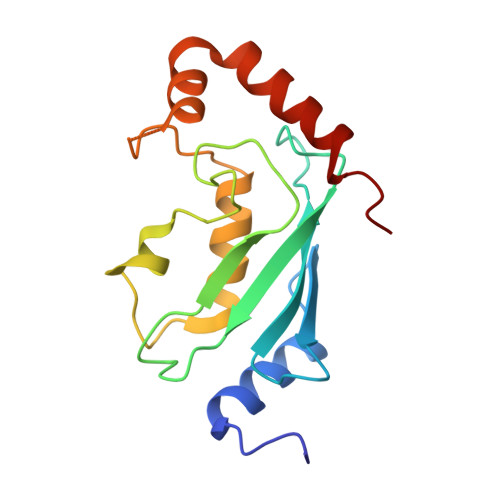
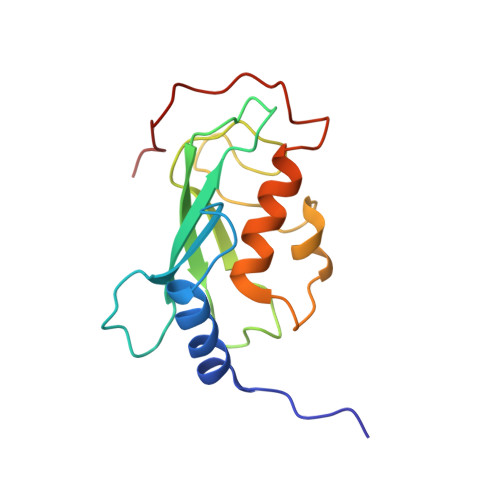
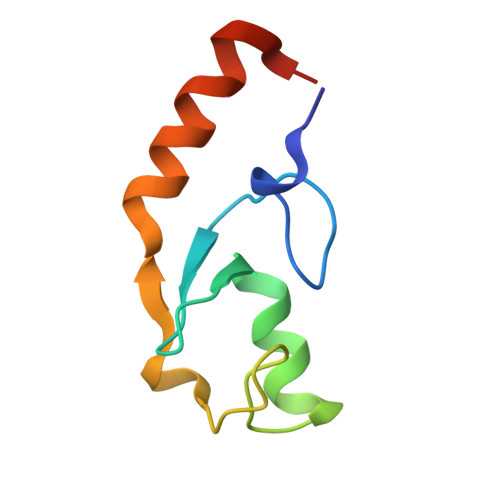
Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex.
Zhang, M., Windheim, M., Roe, S.M., Peggie, M., Cohen, P., Prodromou, C., Pearl, L.H.(2005) Mol Cell 20: 525-538
- PubMed: 16307917
- DOI: https://doi.org/10.1016/j.molcel.2005.09.023
- Primary Citation of Related Structures:
2C2L, 2C2V - PubMed Abstract:
CHIP is a dimeric U box E3 ubiquitin ligase that binds Hsp90 and/or Hsp70 via its TPR-domain, facilitating ubiquitylation of chaperone bound client proteins. We have determined the crystal structure of CHIP bound to an Hsp90 C-terminal decapeptide. The structure explains how CHIP associates with either chaperone type and reveals an unusual asymmetric homodimer in which the protomers adopt radically different conformations. Additionally, we identified CHIP as a functional partner of Ubc13-Uev1a in formation of Lys63-linked polyubiquitin chains, extending CHIP's roles into ubiquitin regulation as well as targeted destruction. The structure of Ubc13-Uev1a bound to the CHIP U box domain defines the basis for selective cooperation of CHIP with specific ubiquitin-conjugating enzymes. Remarkably, the asymmetric arrangement of the TPR domains in the CHIP dimer occludes one Ubc binding site, so that CHIP operates with half-of-sites activity, providing an elegant means for coupling a dimeric chaperone to a single ubiquitylation system.
- Section of Structural Biology, Institute of Cancer Research, Chester Beatty Laboratories, 237 Fulham Road, London SW3 6JB, United Kingdom.
Organizational Affiliation: