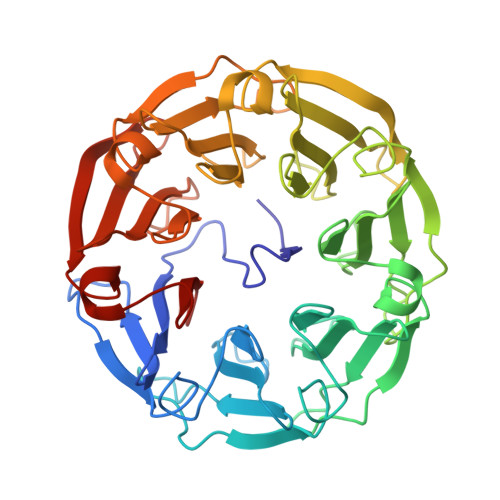
Beta-Propeller Crystal Structure of Psathyrella Velutina Lectin: An Integrin-Like Fungal Protein Interacting with Monosaccharides and Calcium.
Cioci, G., Mitchell, E.P., Chazalet, V., Debray, H., Oscarson, S., Lahmann, M., Gautier, C., Breton, C., Perez, S., Imberty, A.(2006) J Mol Biology 357: 1575
- PubMed: 16497330
- DOI: https://doi.org/10.1016/j.jmb.2006.01.066
- Primary Citation of Related Structures:
2BWM, 2BWR, 2C25, 2C4D - PubMed Abstract:
The lectin from the mushroom Psathyrella velutina recognises specifically N-acetylglucosamine and N-acetylneuraminic acid containing glycans. The crystal structure of the 401 amino acid residue lectin shows that it adopts a very regular seven-bladed beta-propeller fold with the N-terminal region tucked into the central cavity around the pseudo 7-fold axis. In the complex with N-acetylglucosamine, six monosaccharides are bound in pockets located between two consecutive propeller blades. Due to the repeats shown by the sequence the binding sites are very similar. Five hydrogen bonds between the protein and the sugar hydroxyl and N-acetyl groups stabilize the complex, together with the hydrophobic interactions with a conserved tyrosine and histidine. The complex with N-acetylneuraminic acid shows molecular mimicry with the same hydrogen bond network, but with different orientations of the carbohydrate ring in the binding site. The beta-hairpin loops connecting the two inner beta-strands of each blade are metal binding sites and two to three calcium ions were located in the structure. The multispecificity and high multivalency of this mushroom lectin, combined with its similarity to the extracellular domain of an important class of cell adhesion molecules, integrins, are another example of the outstanding success of beta-propeller structures as molecular binding machines in nature.
- Centre de Recherches sur les Macromolécules Végétales (affiliated with Université Joseph Fourier), CNRS, F-38041 Grenoble, France.
Organizational Affiliation: