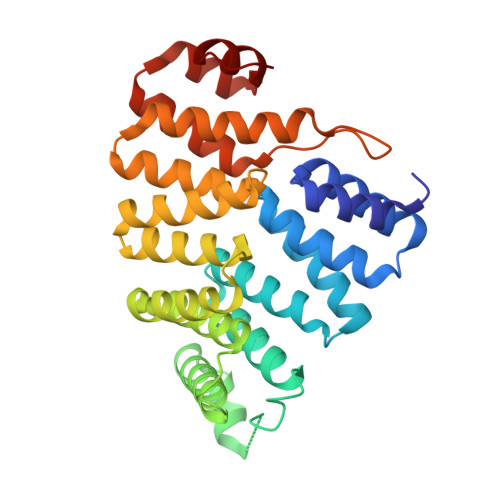
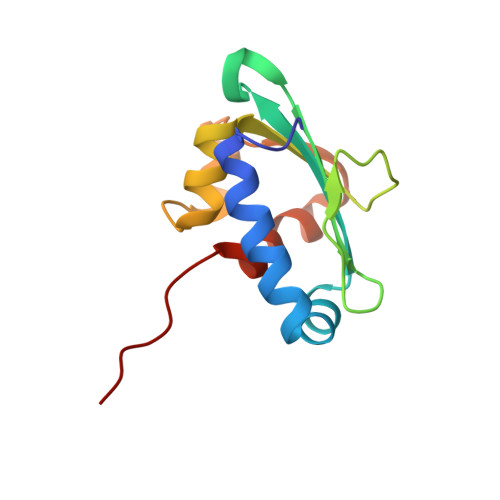
Recognition of a Functional Peroxisome Type 1 Target by the Dynamic Import Receptor Pex5P.
Stanley, W.A., Filipp, F.V., Kursula, P., Schuller, N., Erdmann, R., Schliebs, W., Sattler, M., Wilmanns, M.(2006) Mol Cell 24: 653
- PubMed: 17157249
- DOI: https://doi.org/10.1016/j.molcel.2006.10.024
- Primary Citation of Related Structures:
2C0L, 2C0M - PubMed Abstract:
Peroxisomes require the translocation of folded and functional target proteins of various sizes across the peroxisomal membrane. We have investigated the structure and function of the principal import receptor Pex5p, which recognizes targets bearing a C-terminal peroxisomal targeting signal type 1. Crystal structures of the receptor in the presence and absence of a peroxisomal target, sterol carrier protein 2, reveal major structural changes from an open, snail-like conformation into a closed, circular conformation. These changes are caused by a long loop C terminal to the 7-fold tetratricopeptide repeat segments. Mutations in residues of this loop lead to defects in peroxisomal import in human fibroblasts. The structure of the receptor/cargo complex demonstrates that the primary receptor-binding site of the cargo is structurally and topologically autonomous, enabling the cargo to retain its native structure and function.
- European Molecular Biology Laboratory-Hamburg Outstation, Notkestrasse 85, 22603 Hamburg.
Organizational Affiliation: