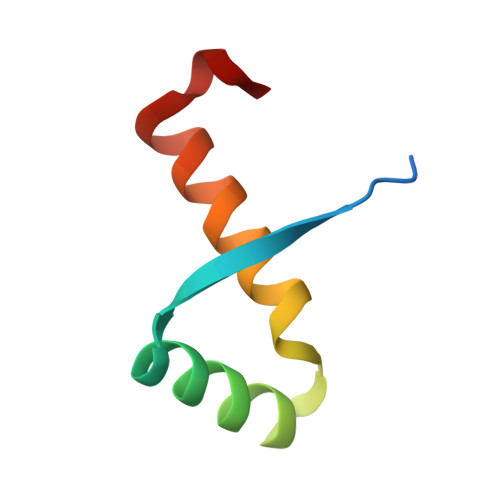
Structures of Omega Repressors Bound to Direct and Inverted DNA Repeats Explain Modulation of Transcription.
Weihofen, W.A., Cicek, A., Pratto, F., Alonso, J.C., Saenger, W.(2006) Nucleic Acids Res 34: 1450
- PubMed: 16528102
- DOI: https://doi.org/10.1093/nar/gkl015
- Primary Citation of Related Structures:
2BNW, 2BNZ, 2CAX - PubMed Abstract:
Repressor omega regulates transcription of genes required for copy number control, accurate segregation and stable maintenance of inc18 plasmids hosted by Gram-positive bacteria. omega belongs to homodimeric ribbon-helix-helix (RHH2) repressors typified by a central, antiparallel beta-sheet for DNA major groove binding. Homodimeric omega2 binds cooperatively to promotors with 7 to 10 consecutive non-palindromic DNA heptad repeats (5'-(A)/(T)ATCAC(A)/(T)-3', symbolized by -->) in palindromic inverted, converging (--><--) or diverging (<---->) orientation and also, unique to omega2 and contrasting other RHH2 repressors, to non-palindromic direct (-->-->) repeats. Here we investigate with crystal structures how omega2 binds specifically to heptads in minimal operators with (-->-->) and (--><--) repeats. Since the pseudo-2-fold axis relating the monomers in omega(2) passes the central C-G base pair of each heptad with approximately 0.3 A downstream offset, the separation between the pseudo-2-fold axes is exactly 7 bp in (-->-->), approximately 0.6 A shorter in (--><--) but would be approximately 0.6 A longer in (<---->). These variations grade interactions between adjacent omega2 and explain modulations in cooperative binding affinity of omega2 to operators with different heptad orientations.
- Institut für Chemie und Biochemie/Kristallographie, Freie Universität Berlin, Takustr. 6, 14195 Berlin, Germany.
Organizational Affiliation: