Structural, Physical, and Biological Characteristics of RNA.DNA Binding Agent N8-Actinomycin D.
Shinomiya, M., Chu, W., Carlson, R.G., Weaver, R.F., Takusagawa, F.(1995) Biochemistry 34: 8481
- PubMed: 7541244
- Primary Citation of Related Structures:
209D - PubMed Abstract:
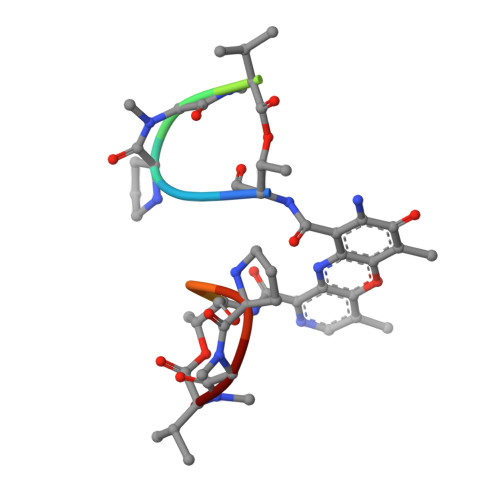
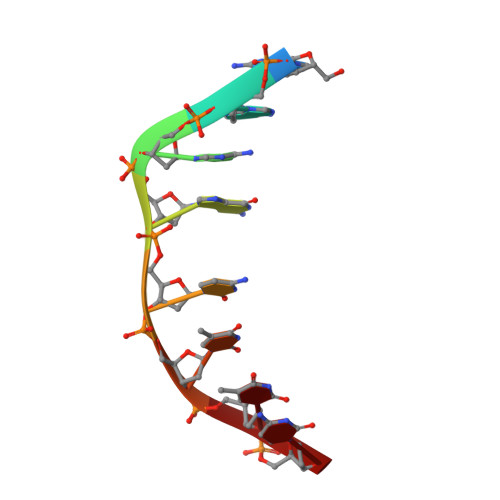
The crystal structure of the self-complementary DNA octamer d(GAAGCTTC)2 complexed with N8-actinomycin D (N8AMD) has been determined at 3.0 A resolution (space group: P3(1)21; unit cell: a = 62.30, b = 62.30, c = 42.97 A; R = 0.173 for 1845 reflections). The DNA structure was severely distorted by the N8AMD bound intercalatively into the middle dinucleotide, 5'-GC-3'. The two cyclic depsipeptides, which differ from each other in overall conformation, lie in the minor groove. The complex is further stabilized by forming base--peptide and chromophore--backbone hydrogen bonds. The complexes are stacked together to form a pseudocontinuous helix running through the crystals. The structure of d(GAAGCTTC)2-actinomycin D (AMD) crystallized in the space group C2 [Kamitori S., & Takusagawa, F. (1992) J. Mol. Biol. 225, 445-456] was re-refined in order to compare it directly to the N8AMD complex structure. The asymmetrical binding mode of AMD has been confirmed on the basis of the two complex structures. The crystal structures of the N8AMD and AMD complexes bound to the same d(GAAGCTTC)2 differed by a root-mean-square deviation on all atom positions of 1.77 A, but most of the structural differences can be attributed to molecular packing in two different crystal forms, and not to structural differences induced by the interaction with the intercalating agents. However, the DNA binding and biological characteristics of N8AMD and AMD are quite different from each other. The DNA association constant of N8AMD is 33-fold less than that of AMD in an aqueous solution. N8AMD required a concentration > 10.0 microM to inhibit RNA synthesis activity in HeLa cells by 50%, whereas AMD reached to the same inhibitory level at only 35 nM. The structure of the DNA-N8AMD complex suggested that substitution of the N-methyl-L-valine residue in the cyclic depsipeptide with a N-methyl-D-valine residue might increase the hydrophobic interaction with the minor groove of the DNA. Thus the DNA association constant and RNA synthesis inhibitory activities of 5,5'-N-methyl-D-valine AMD (D-MeVal-AMD) have also been determined. The DNA association constant of D-MeVal-AMD is more than 2-fold greater than that of AMD, and the RNA synthesis inhibitory activity is about 20-fold greater.
- Departments of Chemistry and Biochemistry, University of Kansas, Lawrence 66045-0046, USA.
Organizational Affiliation: