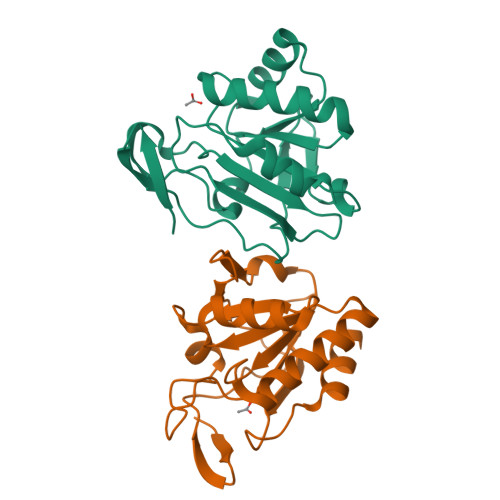
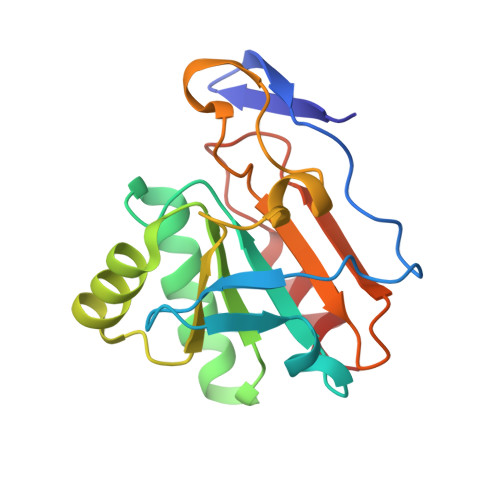
Structure of the inactive variant C60S of Mycobacterium tuberculosis thiol peroxidase
Stehr, M., Hecht, H.J., Jager, T., Flohe, L., Singh, M.(2006) Acta Crystallogr D Biol Crystallogr 62: 563-567
- PubMed: 16627951
- DOI: https://doi.org/10.1107/S0907444906008249
- Primary Citation of Related Structures:
1Y25 - PubMed Abstract:
The genome of Mycobacterium tuberculosis encodes several peroxiredoxins (Prxs) thought to be active against organic and inorganic peroxides. The open reading frame Rv1932 encodes a 165-residue thiol peroxidase (Tpx), which belongs to the atypical 2-Cys peroxiredoxin family. The crystal structure of the C60S mutant of M. tuberculosis Tpx (MtTpx) crystallized in space group P3(1)21, with unit-cell parameters a = 106.08, b = 106.08, c = 65.33 A. The structure has been refined to an R value of 17.1% (R(free) = 24.9%) at 2.1 A resolution. MtTpx is structurally homologous to other peroxiredoxins, including the mycobacterial AhpC and AhpE. The inactive MtTpx C60S mutant structure closely resembles the structure of Streptococcus pneumoniae Tpx (SpTpx) and thus represents the reduced enzyme state. The mutated active-site serine is electrostatically linked to Arg130 and hydrogen bonded to Thr57, practically identical to the cysteine in SpTpx. A cocrystallized acetate molecule mimics the position of the substrate and interacts with Ser60, Arg130 and Thr57.
Organizational Affiliation:
Department GNA, GBF - Gesellschaft für Biotechnologische Forschung, Mascheroder Weg 1, D-38124 Braunschweig, Germany.