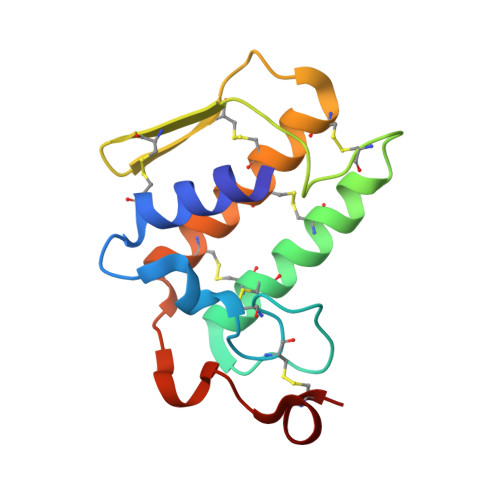
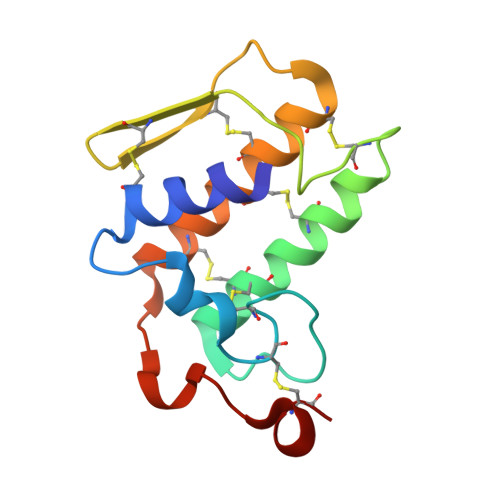
Structure of the zinc-induced heterodimer of two calcium-free isoforms of phospholipase A2 from Naja naja sagittifera at 2.7 angstroms resolution.
Jabeen, T., Sharma, S., Singh, N., Singh, R.K., Verma, A.K., Paramasivam, M., Srinivasan, A., Singh, T.P.(2005) Acta Crystallogr D Biol Crystallogr 61: 302-308
- PubMed: 15735340
- DOI: https://doi.org/10.1107/S0907444904033165
- Primary Citation of Related Structures:
1XXW - PubMed Abstract:
The crystal structure of a zinc-induced heterodimer of two metal-free isoforms of a cobra venom phospholipase A(2) has been determined at 2.7 angstroms resolution. The crystals belong to space group P4(1), with unit-cell parameters a = b = 65.5, c = 58.4 angstroms, and have a single dimer in the asymmetric unit. The structure has been refined to R(cryst) and R(free) factors of 0.188 and 0.232, respectively. The two isoforms have a sequence identity of 82%. The zinc ion forms a fivefold coordination with a trigonal bipyramidal geometry involving one O atom each from Asp24 and Asn112 from molecule A and Asp24 from molecule B and two water molecules. Both molecules of the dimer are inactive. Molecule A is inactive because Arg31 (B) binds to Asp49 (A), while an acetate ion has displaced the essential water molecule and interacts with His48 (A). On the other hand, Arg31 (A) interacts with the calcium-binding loop of molecule B, resulting in an altered conformation of the loop. The absence of a calcium ion, loss of the essential water molecule and the altered conformation of the calcium-binding loop may be the reasons for the loss of activity of molecule B.
- Department of Biophysics, All India Institute of Medical Sciences, New Delhi 110029, India.
Organizational Affiliation: