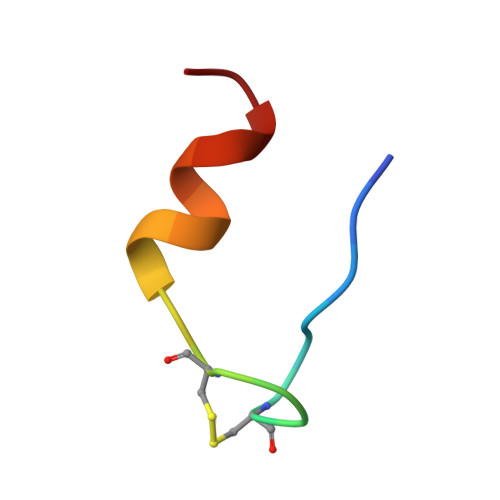
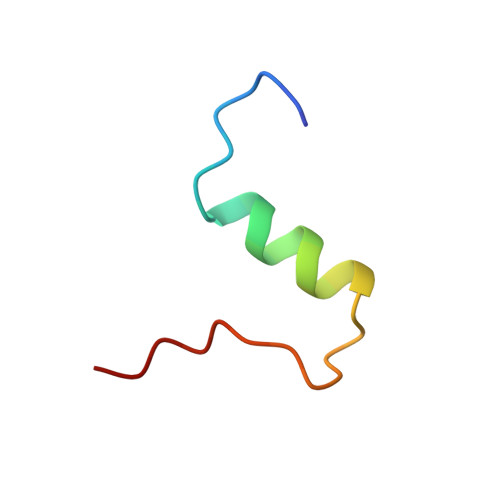
Structure of a protein in a kinetic trap.
Hua, Q.X., Gozani, S.N., Chance, R.E., Hoffmann, J.A., Frank, B.H., Weiss, M.A.(1995) Nat Struct Biol 2: 129-138
- PubMed: 7749917
- DOI: https://doi.org/10.1038/nsb0295-129
- Primary Citation of Related Structures:
1XGL, 2HIU - PubMed Abstract:
We have determined the structure of a metastable disulphide isomer of human insulin. Although not observed for proinsulin folding or insulin-chain recombination, the isomer retains ordered secondary structure and a compact hydrophobic core. Comparison with native insulin reveals a global rearrangement in the orientation of A- and B-chains. One face of the protein's surface is nevertheless in common between native and non-native structures. This face contains receptor-binding determinants, rationalizing the partial biological activity of the isomer. Structures of native and non-native disulphide isomers also define alternative three-dimensional templates. Threading of insulin-like sequences provide an experimental realization of the inverse protein-folding problem.
- Department of Biochemistry and Molecular Biology, University of Chicago, Illinois 60637-5419, USA.
Organizational Affiliation: