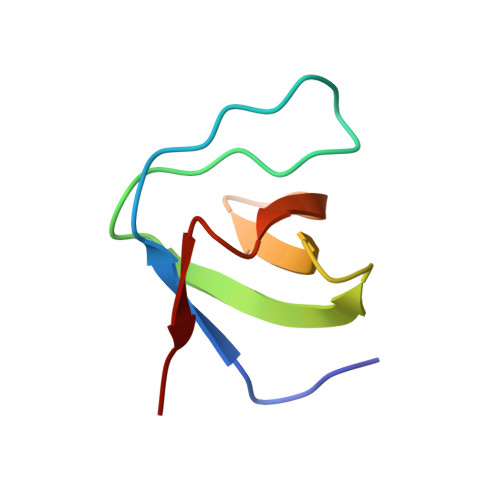
Effects of P47Phox C-Terminus Phosphorylation on Binding Interactions with P40Phox and P67Phox: Structural and Functional Comparison of P40Phox P67Phox SH3 Domains
Massenet, C., Chenavas, S., Cohen-Addad, C., Dagher, M.-C., Brandolin, G., Pebay-Peyroula, E., Fieschi, F.(2005) J Biological Chem 280: 13752
- PubMed: 15657040
- DOI: https://doi.org/10.1074/jbc.M412897200
- Primary Citation of Related Structures:
1W6X, 1W70 - PubMed Abstract:
The neutrophil NADPH oxidase produces superoxide anions in response to infection. This reaction is activated by association of cytosolic factors, p47phox and p67phox, and a small G protein Rac with the membranous flavocytochrome b558. Another cytosolic factor, p40phox, is associated to the complex and is reported to play regulatory roles. Initiation of the NADPH oxidase activation cascade has been reported as consecutive to phosphorylation on serines 359/370 and 379 of the p47phox C terminus. These serines surround a polyproline motif that can interact with the Src homology 3 (SH3) module of p40phox (SH3p40) or the C-terminal SH3 of p67phox (C-SH3p67). The latter one presents a higher affinity in the resting state for p47phox. A change in SH3 binding preference following phosphorylation has been postulated earlier. Here we report the crystal structures of SH3p40 alone or in complex with a 12-residue proline-rich region of p47phox at 1.46 angstrom resolution. Using intrinsic tryptophan fluorescence measurements, we compared the affinity of the strict polyproline motif and the whole C terminus peptide with both SH3p40 and C-SH3p67. These data reveal that SH3p40 can interact with a consensus polyproline motif but also with a noncanonical motif of the p47phox C terminus. The electrostatic surfaces of both SH3 are very different, and therefore the binding preference for C-SH3p67 can be attributed to the polyproline motif recognition and particularly to the Arg-368p47 binding mode. The noncanonical motif contributes equally to interaction with both SH3. The influence of serine phosphorylation on residues 359/370 and 379 on the affinity for both SH3 domains has been checked. We conclude that contrarily to previous suggestions, phosphorylation of Ser-359/370 does not modify the SH3 binding affinity for both SH3, whereas phosphorylation of Ser-379 has a destabilizing effect on both interactions. Other mechanisms than a phosphorylation induced switch between the two SH3 must therefore take place for NADPH oxidase activation cascade to start.
- Institut de Biologie Structurale, UMR 5075 CEA/CNRS/Université Joseph Fourier, Laboratoire des Protéines Membranaires, 41 rue Jules Horowitz 38027 Grenoble cedex 1, France.
Organizational Affiliation: