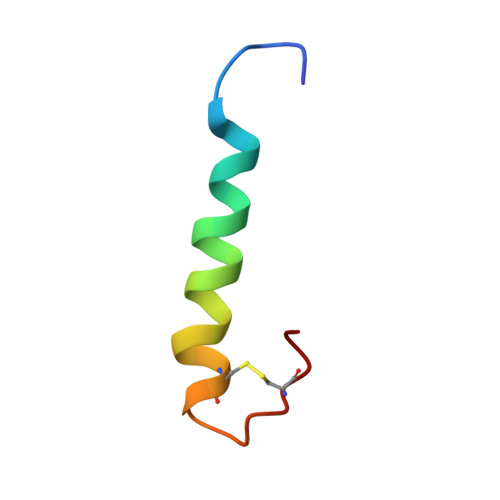
The solution structure of the FATC domain of the protein kinase target of rapamycin suggests a role for redox-dependent structural and cellular stability.
Dames, S.A., Mulet, J.M., Rathgeb-Szabo, K., Hall, M.N., Grzesiek, S.(2005) J Biological Chem 280: 20558-20564
- PubMed: 15772072
- DOI: https://doi.org/10.1074/jbc.M501116200
- Primary Citation of Related Structures:
1W1N - PubMed Abstract:
The target of rapamycin (TOR) is a highly conserved Ser/Thr kinase that plays a central role in the control of cellular growth. TOR has a characteristic multidomain structure. Only the kinase domain has catalytic function; the other domains are assumed to mediate interactions with TOR substrates and regulators. Except for the rapamycin-binding domain, there are no high-resolution structural data available for TOR. Here, we present a structural, biophysical, and mutagenesis study of the extremely conserved COOH-terminal FATC domain. The importance of this domain for TOR function has been highlighted in several publications. We show that the FATC domain, in its oxidized form, exhibits a novel structural motif consisting of an alpha-helix and a COOH-terminal disulfide-bonded loop between two completely conserved cysteine residues. Upon reduction, the flexibility of the loop region increases dramatically. The structural data, the redox potential of the disulfide bridge, and the biochemical data of a cysteine to serine mutant indicate that the intracellular redox potential can affect the cellular amount of the TOR protein via the FATC domain. Because the amount of TOR mRNA is not changed, the redox state of the FATC disulfide bond is probably influencing the degradation of TOR.
- Department of Structural Biology, Biozentrum, University of Basel, Klingelbergstrasse 70, 4056 Basel, Switzerland. sonja.dames@unibas.ch
Organizational Affiliation: