Thermodynamic and Structural Equivalence of Two Hla-B27 Subtypes Complexed with a Self-Peptide
Hulsmeyer, M., Welfle, K., Pohlmann, T., Misselwitz, R., Alexiev, U., Welfle, H., Saenger, W., Uchanska-Ziegler, B., Ziegler, A.(2005) J Mol Biology 346: 1367
- PubMed: 15713487
- DOI: https://doi.org/10.1016/j.jmb.2004.12.047
- Primary Citation of Related Structures:
1W0V, 1W0W - PubMed Abstract:
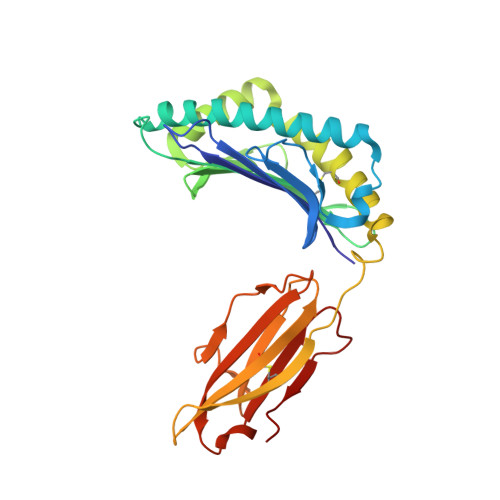
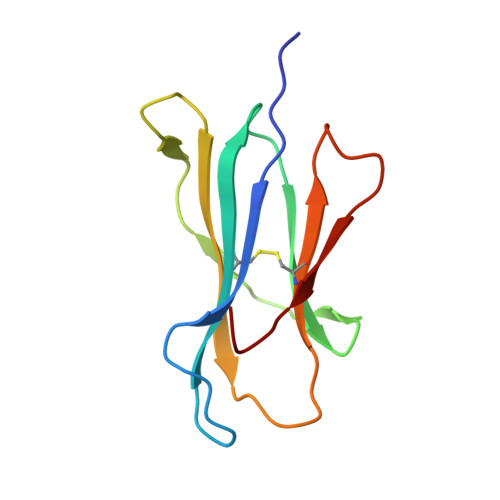
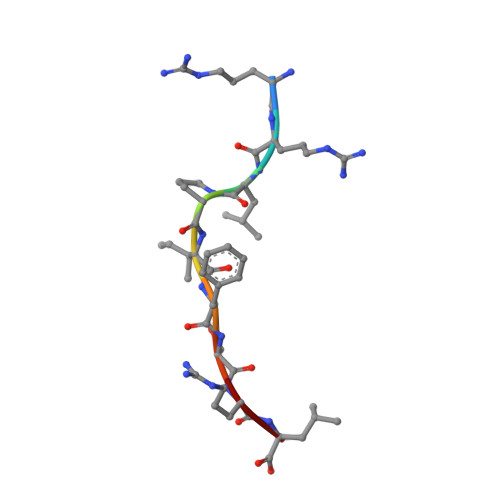
The F pocket of major histocompatibility complex (in humans HLA) class I molecules accommodates the C terminus of the bound peptide. Residues forming this pocket exhibit considerable polymorphism, and a single difference (Asp116 in HLA-B*2705 and His116 in HLA-B*2709 heavy chains) confers differential association of these two HLA-B27 subtypes to the autoimmune disease ankylosing spondylitis. As peptide presentation by HLA molecules is of central importance for immune responses, we performed thermodynamic (circular dichroism, differential scanning calorimetry, fluorescence polarization) and X-ray crystallographic analyses of both HLA-B27 subtypes complexed with the epidermal growth factor response factor 1-derived self-peptide TIS (RRLPIFSRL) to understand the impact of the Asp116His exchange on peptide display. This peptide is known to be presented in vivo by both subtypes, and as expected for a self-peptide, TIS-reactive cytotoxic T lymphocytes are absent in the respective individuals. The thermodynamic analyses reveal that both HLA-B27:TIS complexes exhibit comparable, relatively high thermostability (Tm approximately 60 degrees C) and undergo multi-step unfolding reactions, with dissociation of the peptide in the first step. As shown by X-ray crystallography, only subtle structural differences between the subtypes were observed regarding the architecture of their F pockets, including the presence of distinct networks of water molecules. However, no consistent structural differences were found between the peptide presentation modes. In contrast to other peptides displayed by the two HLA-subtypes which show either structural or dynamical differences in their peptide presentation modes, the TIS-complexed HLA-B*2705 and HLA-B*2709 subtypes are an example for thermodynamic and structural equivalence, in agreement with functional data.
- Institut für Chemie/Kristallographie, Freie Universität Berlin, Takustrasse 6, 14195 Berlin, Germany.
Organizational Affiliation: