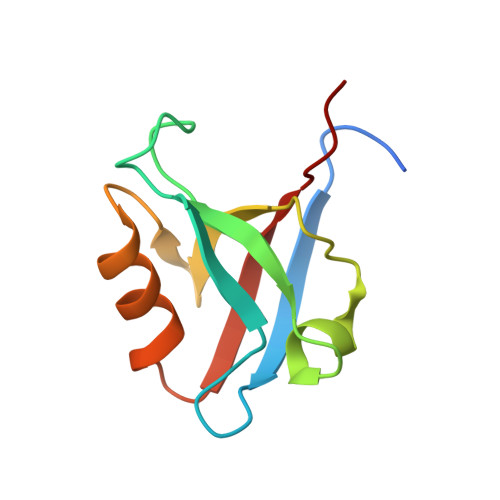
Demonstration of long-range interactions in a PDZ domain by NMR, kinetics, and protein engineering.
Gianni, S., Walma, T., Arcovito, A., Calosci, N., Bellelli, A., Engstrom, A., Travaglini-Allocatelli, C., Brunori, M., Jemth, P., Vuister, G.W.(2006) Structure 14: 1801-1809
- PubMed: 17161370
- DOI: https://doi.org/10.1016/j.str.2006.10.010
- Primary Citation of Related Structures:
1VJ6 - PubMed Abstract:
Understanding the basis of communication within protein domains is a major challenge in structural biology. We present structural and dynamical evidence for allosteric effects in a PDZ domain, PDZ2 from the tyrosine phosphatase PTP-BL, upon binding to a target peptide. The NMR structures of its free and peptide-bound states differ in the orientation of helix alpha2 with respect to the remainder of the molecule, concomitant with a readjustment of the hydrophobic core. Using an ultrafast mixing instrument, we detected a deviation from simple bimolecular kinetics for the association with peptide that is consistent with a rate-limiting conformational change in the protein (k(obs) approximately 7 x 10(3) s(-1)) and an induced-fit model. Furthermore, the binding kinetics of 15 mutants revealed that binding is regulated by long-range interactions, which can be correlated with the structural rearrangements resulting from peptide binding. The homologous protein PSD-95 PDZ3 did not display a similar ligand-induced conformational change.
- Istituto di Biologia e Patologia Molecolari del CNR, Dipartimento di Scienze Biochimiche A. Rossi Fanelli, Università di Roma La Sapienza, Piazzale A. Moro 5, 00185 Rome, Italy.
Organizational Affiliation: