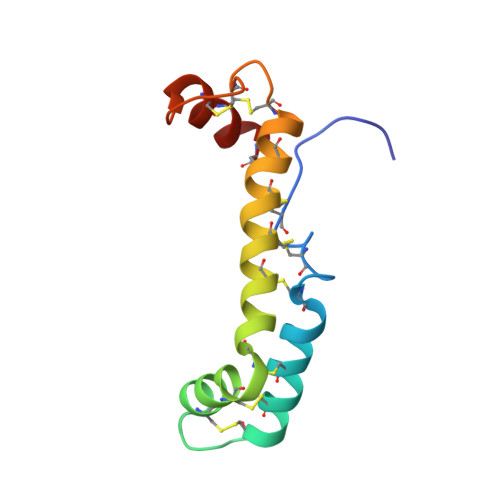
Solution Structure of Vanabin2, a Vanadium(IV)-Binding Protein from the Vanadium-Rich Ascidian Ascidia sydneiensis samea
Hamada, T., Asanuma, M., Ueki, T., Hayashi, F., Kobayashi, N., Yokoyama, S., Michibata, H., Hirota, H.(2005) J Am Chem Soc 127: 4216-4222
- PubMed: 15783203
- DOI: https://doi.org/10.1021/ja042687j
- Primary Citation of Related Structures:
1VFI - PubMed Abstract:
Ascidians belonging to the suborder Phlebobranchia are known to accumulate high levels of a transition metal, vanadium, in their blood cells, called vanadocytes, although the mechanism for this biological phenomenon remains unclear. Recently, we identified vanadium(IV)-binding proteins, designated as Vanabins, from vanadium-accumulating ascidians. Here, we report the first 3D structure of Vanabin2 from an ascidian, Ascidia sydneiensis samea, in an aqueous solution. The structure revealed a novel bow-shaped conformation, with four alpha-helices connected by nine disulfide bonds. There are no structural homologues reported so far. The 15N heteronuclear single-quantum coherence (HSQC) perturbation experiments of Vanabin2 indicated that vanadyl cations, which are exclusively localized on the same face of the molecule, are coordinated by amine nitrogens derived from amino acid residues such as lysines, arginines, and histidines, as suggested by the electron paramagnetic resonance (EPR) results. The present NMR studies provide information that will contribute toward elucidating the mechanism of vanadium accumulation in ascidians.
- RIKEN Genomic Sciences Center, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama 230-0045, Japan.
Organizational Affiliation: